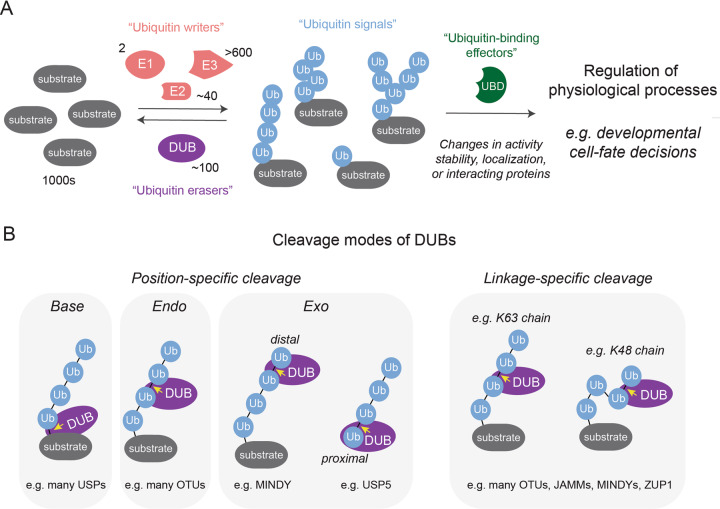
Fig. 1. Overview of how ubiquitin signaling regulates developmental cell-fate decisions and cleavage modes of DUBs.
A To initiate ubiquitin signaling, an enzymatic cascade, consisting of ubiquitin E1 activating, E2 conjugating, and E3 ligating enzymes, decorates substrates with topologically different ubiquitin modifications. Effector proteins containing various ubiquitin-binding domains (UBDs) interpret the ubiquitin signals and mediate changes in the substrate activity, stability, localization or interacting proteins. This controls cellular behavior during many physiological processes, including development. DUBs are important regulators of this ubiquitin code by reversing ubiquitin modifications, thus modulating or terminating signaling. B Cartoons depicting different position- and linkage-specific cleavage modes by which DUBs can act on their substrates. The yellow arrow indicates the peptide/isopeptide bond that is hydrolyzed in each example.