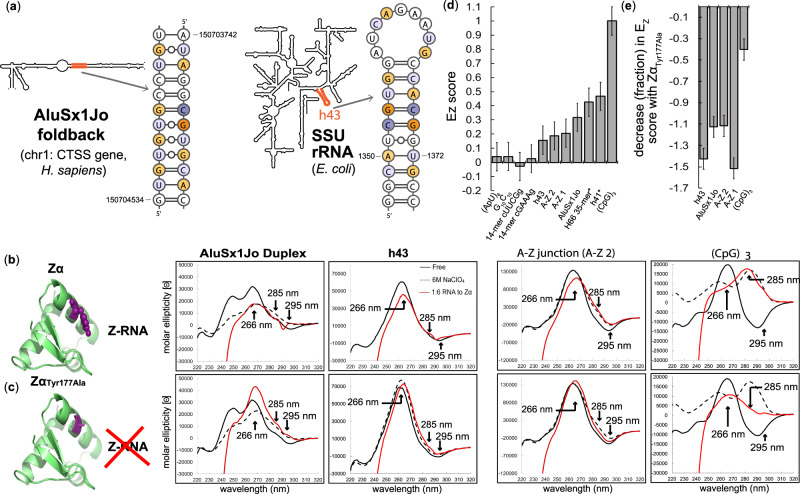
Fig. 2. Zα induces a partial Z-conformation in AluSx1Jo and h43.
a Location of AluSx1Jo and h43 on secondary structure diagrams of the CTSS gene (Chromosome 1) from H. sapiens and the small subunit ribosomal RNA from E. coli. For h43, the first three G = C base pairs were engineered for added stability. In this and subsequent figures, CpG and YpR steps are shown in dark (CpG) and light (non-CpG YpR) shades of purple/orange colors. b CD spectra of the AluSx1Jo and h43 RNAs in absence of protein (black) and at the 1:6 RNA:Zα ratio (red), at which binding is saturated. Controls: 6 M NaClO4 (dotted line), ionic condition that promotes Z-RNA formation of CpG repeats57; A-Z junction35 (second to right-most panel), which is an RNA that has a (CpG)6 sequence followed by an A-RNA forming sequence, and (CpG)3 (right-most panel), which is fully converted to Z-RNA at a 1:2 RNA:Zα ratio. c Same as b, but with ZαTyr177Ala instead of wild-type Zα. d EZ scores quantifying the extent of Z-conformation for the following fragments: (ApU)6, G10C10, 14mer cUUCGg, cGAAAg tetraloop (negative controls); h43 from E. coli; AluSx1Jo; H66 35mer from H. sapiens; (CpG)3 (positive control). e Reduction in EZ score for AluSx1Jo, h43, and (CpG)3 RNAs (expressed as a fraction) when ZαTyr177Ala is used instead of Zα. * indicates that RNA forms a duplex as determined by AUC when stem-loop was expected (Supplementary Fig. 5). An error of 0.1 was determined to be appropriate for the calculated EZ scores by taking into account the difference between the (CpG)3 and (CpG)6 RNAs (which theoretically both have an EZ score of 1) and the difference in the EZ score between repeat measurements on h43 E.coli. All other EZ scores were determined from one CD measurement.