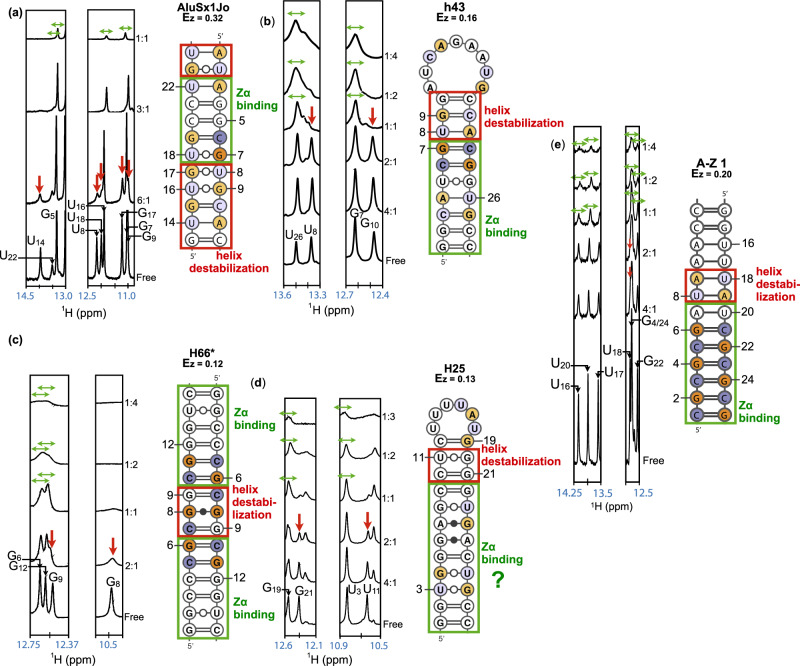
Fig. 3. Pinpointing Zα binding sites and helix destabilization within various RNA sequence contexts.
1H-1D spectra of the following fragments: a AluSx1Jo, b h43 from E. coli, c H66* (extended duplex) from H. sapiens, d H25 from H. sapiens, e A-Z 1, at decreasing RNA:Zα ratios (right-hand side of the spectra slices). Peak disappearance (vertical red arrows) at low concentrations of Zα (below 1:1) caused by destabilization of the A-form helix required for A-Z junction formation is shown. Further line broadening at higher stoichiometric ratios of Zα to RNA (1:1, 1:2, 1:3, 1:4 RNA:Zα) indicate further binding of Zα proteins and growing complex size (green horizontal arrows). Full imino spectra are shown in Supplementary Fig. 8. For H25, due to the lack of information on the imino protons of G6 and G24, in addition to being unable to determine whether G4 is destabilized or coalesces with G28, the exact Zα binding region cannot be confirmed. NMR measurements were performed once as customary, and showed consistency with CD, ITC, and AUC experiments.