Figure 1. scRNA‐seq captures the expression profiles of effectors genes in the late stages of Drosophila embryogenesis.
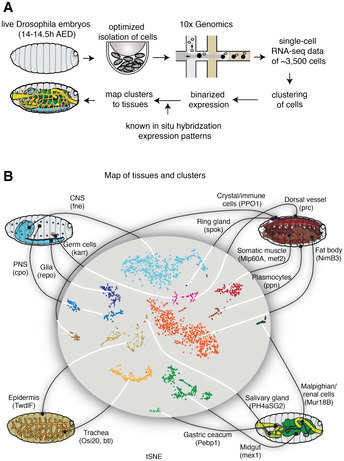
- Single cells were isolated from Drosophila embryos 14–14.5 h after egg deposition (AED). Isolated cells were processed through a 10× Genomics instrument. After sequencing the resulting libraries, the reads were aligned and processed using the standard pipeline from 10× Genomics. The single‐cell gene expression profiles were used to map the cells to known cell types by comparing against the available in situ hybridization patterns from the Berkeley Drosophila Genome Project.
- A tSNE projection of the scRNA‐seq data is shown in the middle, and the known tissues to which the clusters were assigned to are graphically illustrated outside. Marker genes for each tissue type are shown in parentheses.
