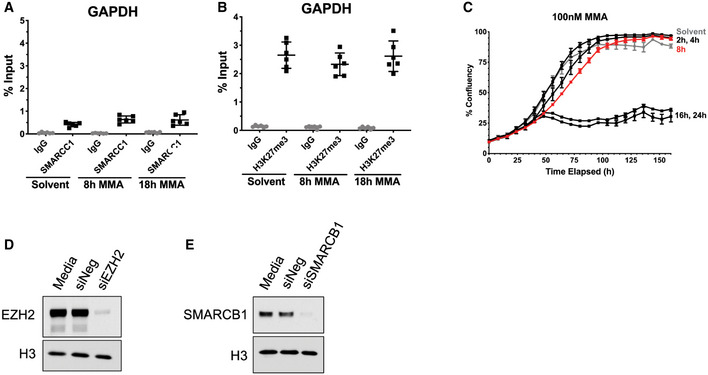
Figure EV2. Mithramycin sensitivity in rhabdoid tumor is not due to reprogramming of housekeeping genes or incomplete loss of siRNA knockdown.
-
A, BChromatin immunoprecipitation of IgG, SMARCC1 (A), or H3K27me3 (B) at the control locus, GAPDH. Data represent mean with standard deviation derived from three independent experiments.
-
CTime course of 100 nM mithramycin exposure in G401 cells. Cells were treated with 100 nM MMA for the indicated times followed by a replacement of drug‐free media. After 8 h (red) of mithramycin exposure, the cells have an irreversible suppression of proliferation compared to solvent control. Data from A, B represent mean with standard deviation derived from three independent experiments. Data in C are mean with standard deviation of 3 biological replicates and representative of three independent experiments.
-
D, EWestern blot confirming efficient protein knockdown of EZH2 in BT12 rhabdoid tumor cells (D) and SMARCB1 in U2OS osteosarcoma cells (E) compared with media and siNegative controls indicative of silencing in Fig 4E and F.
Source data are available online for this figure.
