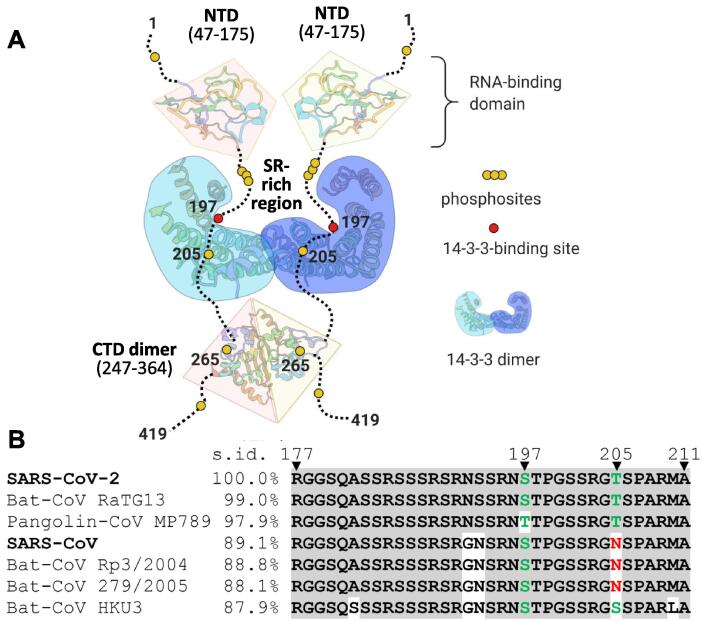
Figure 9.
Association of SARS-CoV-2 N with human 14-3-3. A. A topology model for the complex of SARS-CoV-2 N protein dimer with the dimer of human 14-3-3 illustrating the occlusion of the SR-rich region. Although pSer197 is critical for the 14-3-3 binding, other phosphosites (e.g., the semiconserved Thr205) may play a secondary role. B. Local alignment of SR-rich regions of the most similar coronavirus N proteins in order of descending sequence identity (s.id.) determined using entire N protein sequences. Alignment was performed using Clustal omega and visualized using Mview (https://www.ebi.ac.uk/Tools/msa/mview/) with the following coloring scheme. Residues identical to those in the SARS-CoV-2 sequence are shadowed by grey, phosphorylatable residues in positions 197 and 205 are in green color, residues blocking phosphorylation in position 205 are in red.