Figure 3.
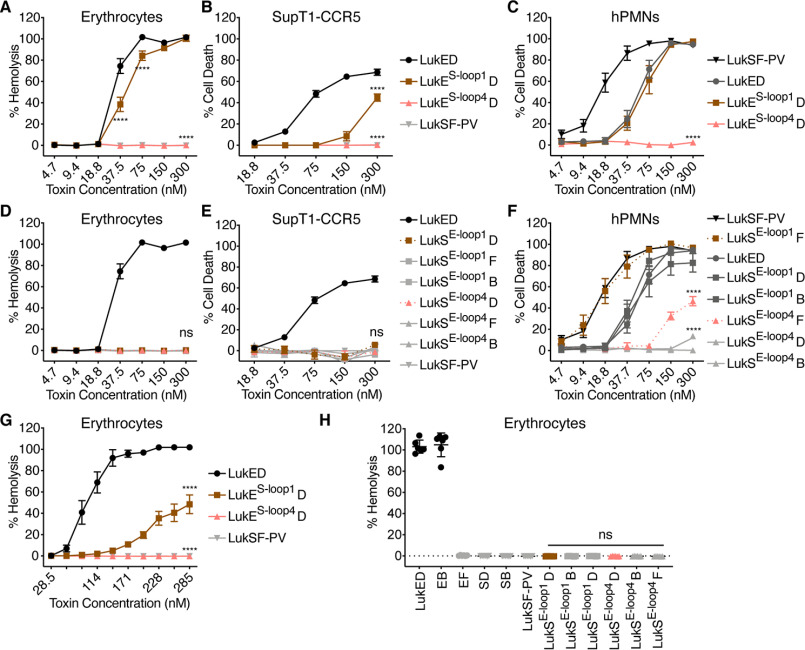
Amino acids 249–253 of LukE are not sufficient for erythrocyte targeting. A and D, hemolysis of primary human erythrocytes treated with LukE and LukS loop chimeras and indicated F subunits (n = 6 donors for erythrocytes). B and E, viability of CCR5 expressing SupT1 cells (SupT1-CCR5) treated with LukE and LukS loop chimeras and indicated F subunits (n = 3–4 experiments). C and F, viability of primary human PMNs treated with LukE and LukS loop chimeras and indicated F subunits (n = 6 donors). G and H, hemolysis of primary human erythrocytes treated with LukE and LukS loop chimeras, in PBS as in Ref. 16 (n = 6 donors). In H, a concentration of 1430 nm per subunit was used, as in Ref. 16. The legend in B applies to A and B, the legend in E applies to D and E. LukES-loop1 indicates LukE with residues 65–70 of LukS-PV, LukES-loop4 indicates LukE with residues 249–253 of LukS-PV, LukSE-1oop1 indicates LukS-PV with residues 65–70 of LukE, and LukSE-loop4 indicates LukS-PV with residues 237–271 of LukE. D indicates LukD, F indicates LukF-PV, and B indicates HlgB. Noncanonical toxin pairs are denoted as EB for LukE + HlgB, etc. See Fig. S1 for amino acid sequences and Fig. S2 (A and C) for cartoon and gel of S subunit loop chimeras. The data are pooled from at least two independent experiments and represent means ± S.E., mean ± S.D. for H. ****, p < 0.0001; ns, not significant. A–G, two-way ANOVA with Sidak's correction, compared with WT LukE (A–C and G) or WT LukSF-PV (D–F). H, one-way ANOVA with Tukey's correction, compared with WT LukSF-PV.