Figure 7.
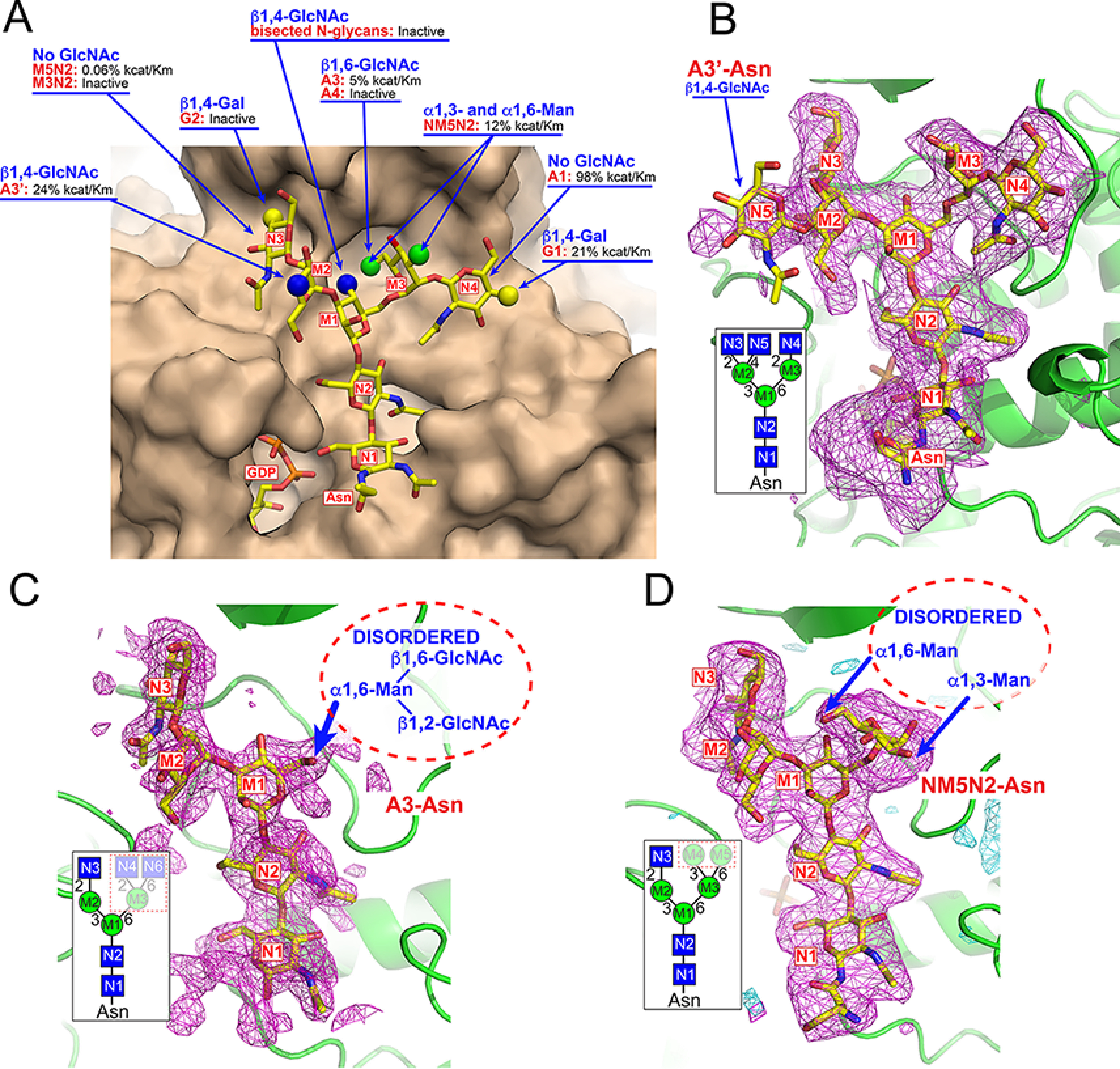
Impact of acceptor modifications on enzyme activity and structures of acceptor complexes. Modifications to the acceptor glycan structure impacts catalytic efficiency in enzyme assays (A). Surface representation of the FUT8:A2-Asn complex (tan) with the A2-Asn structure are displayed in yellow stick representation. Alternative acceptor structures are indicated by spherical representations at positions where they differ from the A2-Asn structure. For each modification, the respective glycan structure is shown in red text and the effect on enzyme activity is listed based on enzyme assay data from Table S1. In cases where the respective acceptor was listed as “inactive” there was no activity detected in the in vitro enzyme assays. Values for kcat/Km are listed by comparison to activity using the A2-Asn substrate. The lack of activity toward the G2-Asn substrate or the glycans with bisected GlcNAcs were based on prior assays from other groups (73, 75, 82). The Polder maps (115) (magenta mesh and contoured at 2.8 σ and 3.5 σ, respectively) were obtained from the crystal structures of three additional acceptor complexes and are shown in B–D. The Polder maps (115) for A3'-Asn, A3-Asn, and NM5N2-Asn acceptors were calculated following a modified procedure to reduce model bias (see “Experimental procedures”). B, the structure of the FUT8:GDP:A3'-Asn complex was essentially the same as the FUT8:GDP:A2-Asn complex except for the additional β1,4-GlcNAc residue (N5) extending from the Man-α1,3- residue (M2). This residue extends into the solvent and has no additional interactions with the enzyme surface. The inset in B indicates the cartoon representation of the A3'-Asn glycan with residue labeling that is also used to label the residues in the difference density (Polder) map. C, the structure of the FUT8:GDP:A3-Asn complex retained the density for the GlcNAc-β1,2Man-α1,3Man-β1,4GlcNAc-β1,4GlcNAc-βAsn region of the A2-Asn complex. However, the entire extension from the Man-α1,6- arm (residues M3, N4, and N6) was disordered and not resolved in the structure (whited out region in the dotted box for the inset; structure and dotted oval in the Polder map (115)). D, the structure of the FUT8:GDP:NM5N2-Asn complex also retained the electron density for the GlcNAc-β1,2Man-α1,3Man-β1,4GlcNAc-β1,4GlcNAc-βAsn region similar to the A2-Asn complex along with density for the Man-α1,6- (M3) residue extending from the Man-β1,4- residue (M1). However, the additional terminal Man-α1,3- (M4) and Man-α1,6- (M5) residues in the NM5N2-Asn structure were disordered and not resolved in the structure (whited out region in dotted box for the inset structure and dotted oval in the Polder map (115).
