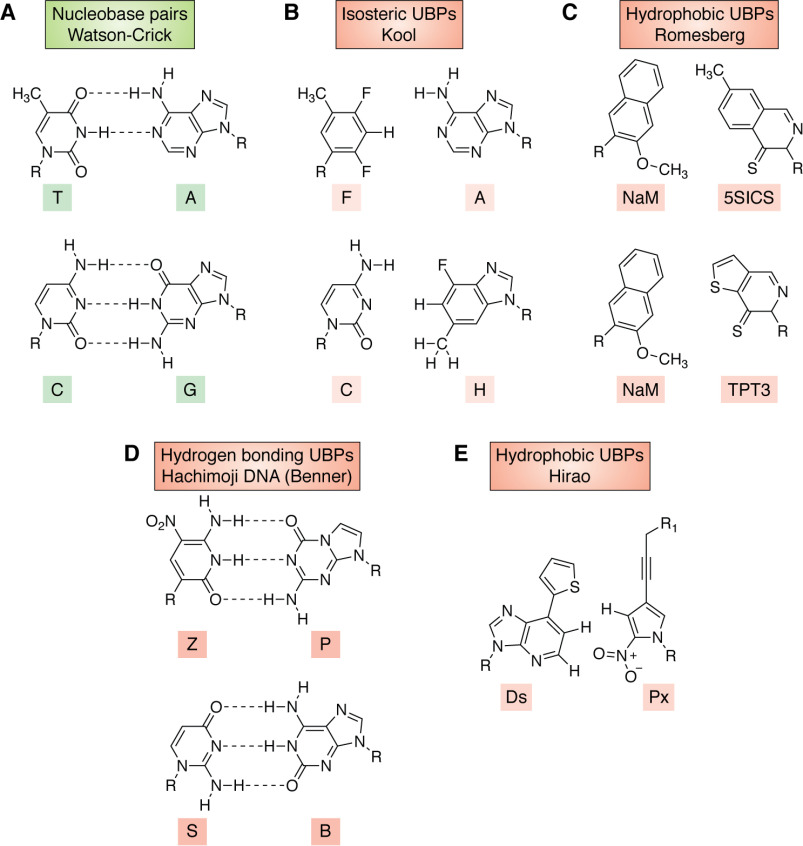
Figure 1.
Chemical structures of nucleobase pairs. R indicates the point of covalent attachment to either deoxyribose or ribose in DNA or RNA, respectively. A, Watson–Crick nucleobase pairs (7). B, isosteric UBPs developed by Kool and co-workers (8). C, hydrophobic UBPs developed by Romesberg and co-workers (9, 10). D, hydrogen-bonding UBPs used in hachimoji DNA developed by Benner and co-workers (11, 12). E, hydrophobic UBPs developed by Hirao and co-workers (13, 14), where R1 can be a variety of substituents.