Figure 4.
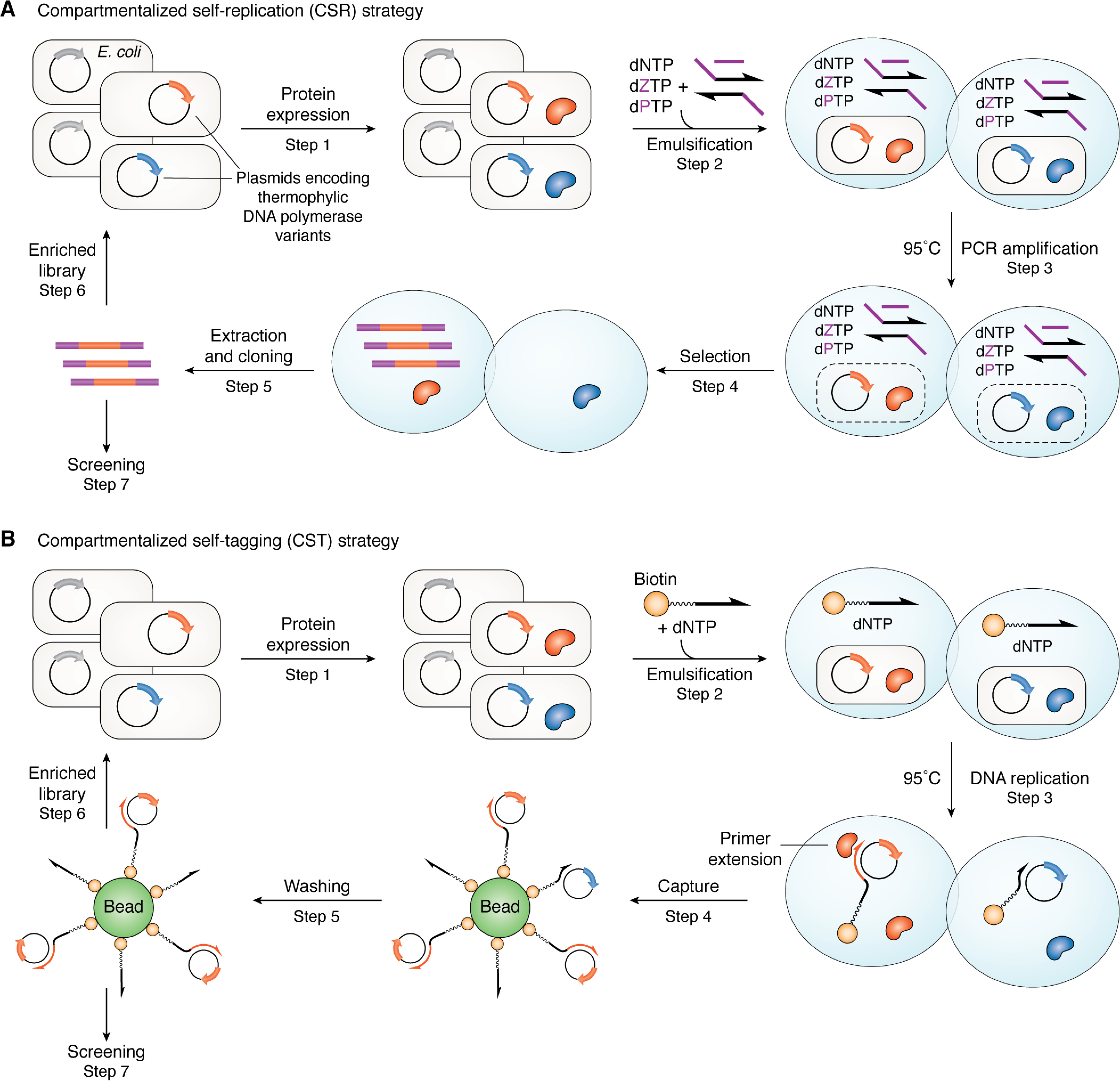
A, overview of a modified CSR strategy for identifying DNA polymerase variants capable of replicating AEGIS UBPs from a library. Each E. coli cell carries a thermophilic DNA polymerase variant (Step 1) and is emulsified with dNTPs and nested primers within a single water droplet. Heating lyses the cells and inactivates endogenous mesophilic polymerases (Step 2). PCR amplification is then performed (Step 3). Polymerases that can replicate UBPs (red) amplify their respective genes, enriching the pool of variant genes in the pool; those that cannot replicate UBPs fail to do so, so their genes are lost (Step 4). The amplified genes encoding polymerase variants that can replicate UBPs are extracted after breaking the emulsion and used to create a new library of plasmids (Step 5) that can be subjected to a further selection round (Step 6). After an appropriate number of selection rounds, the pool of enriched genes can be sequenced (Step 7) and used to express polymerase variants that can be characterized using standard in vitro methods. Based on Fig. 3 of Ref. 65. B, In CST, each E. coli cell carries a thermophilic DNA polymerase variant encoded on a plasmid (Step 1), which also contains a sequence containing one or more UBPs complementary to a single biotin-linked (“tagged”) primer and is emulsified with dNTPs and the tagged primer within a single water droplet. Heating lyses the cells and inactivates endogenous mesophilic polymerases (Step 2). Subsequent DNA replication (Step 3) by the thermophilic DNA polymerase variant results in a “biotin-tagged” form of the plasmid containing the gene (red) encoding the polymerase variant that can replicate the UBP(s). Polymerases that cannot replicate the UBP (blue) will only give weakly stable primer-plasmid complexes. After breaking the emulsion, tagged plasmids are captured onto a bead (Step 4) in proportion to the stability of the extended primer-plasmid complex (Step 5). The recovered plasmid DNA is then amplified and used in a new round of selection (Step 6) or screened (Step 7). Based on Fig. S1 of Ref. 78).
