Figure 2.
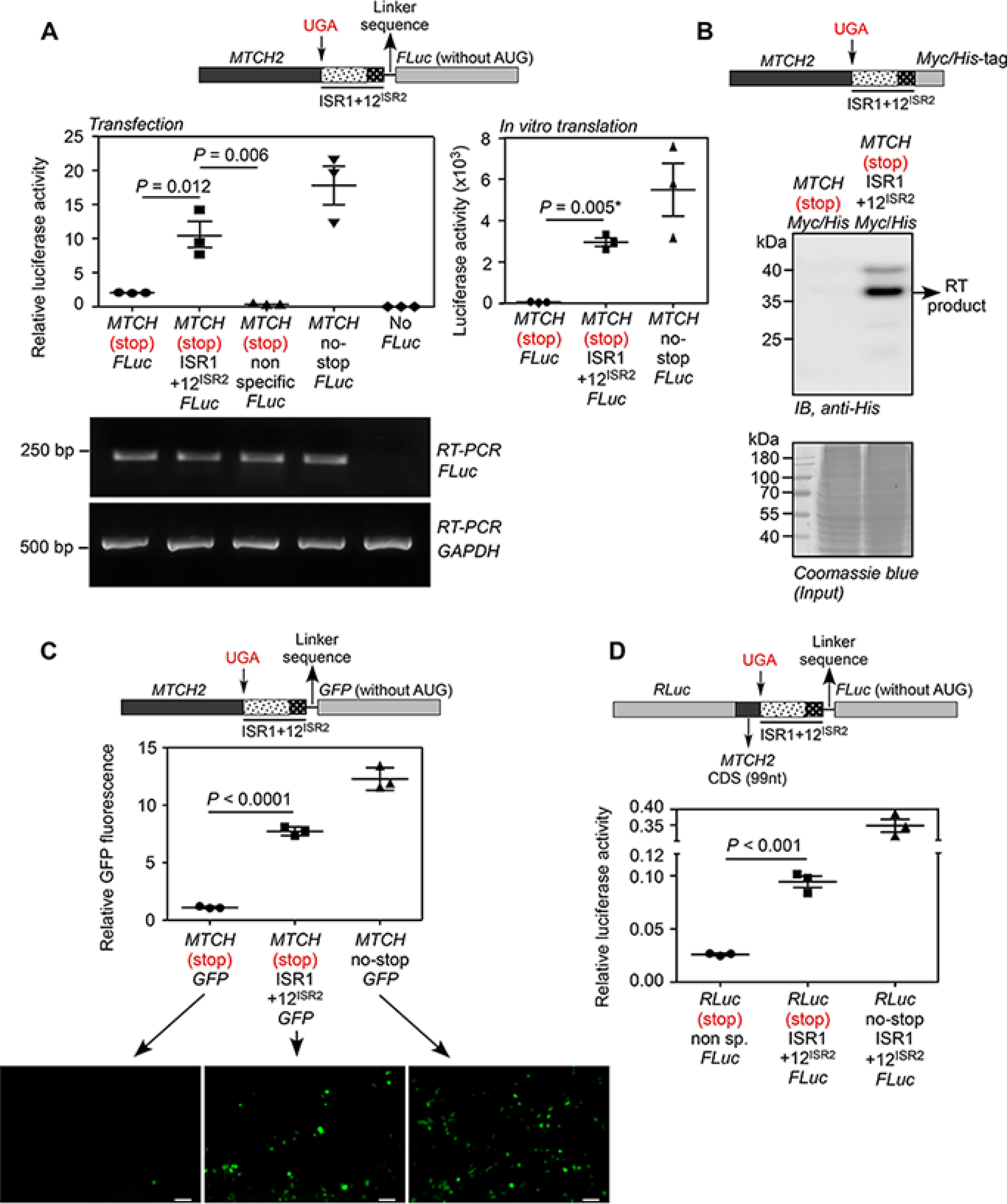
Demonstration of translational read-through across the canonical stop codon of MTCH2. A, luminescence-based assay: plasmid expressing in-frame MTCH2-(UGA)-(ISR1 + 12ISR2)-FLuc and its variants were transfected in HEK293 cells. FLuc activity relative to the activity of co-transfected Renilla luciferase (RLuc) is shown (left). A construct without ISR1 + 12ISR2 between MTCH2 and FLuc, and another construct with a nonspecific sequence between them served as negative controls. RT-PCR of FLuc mRNA is shown. The constructs were subjected to in vitro transcription and in vitro translation using rabbit reticulocyte lysate. Luciferase activity is shown (right). B, Western blotting-based assay: HEK293 cells were transfected with MTCH2-(UGA)-(ISR1 + 12ISR2)-Myc/His construct. His-tagged translational read-through product was enriched using Ni-NTA column and was detected by Western blotting using anti-His antibody. RT, read-through. C, fluorescence-based assay: HEK293 cells were transfected with constructs similar to the one described in A, but the coding sequence of FLuc was replaced with GFP. Fluorescence was analyzed by flow cytometry (graph) and fluorescence microcopy (images below). Scale bar, 50 μm. Graph shows the mean intensity of GFP fluorescence relative to that of co-transfected RFP fluorescence. D, dual luciferase-based assay: 99 nucleotides from the 3′ end of the MTCH2 coding sequence and ISR1 + 12ISR2 were cloned between the coding sequences of RLuc and FLuc (see schematic). All were in-frame. This construct was transfected in HEK293 cells and FLuc activity relative to RLuc is shown. All graphs show mean ± S.E. (n = 3). Results are representatives of at least three independent experiments done in triplicate. Two-tailed Student's t test was used to calculate the p value. *, Welch's correction was applied. MTCH, MTCH2; 12ISR2, first 12 nucleotides of ISR2 (UCCCAGAUGCAC); CDS, coding sequence.
