Figure 4.
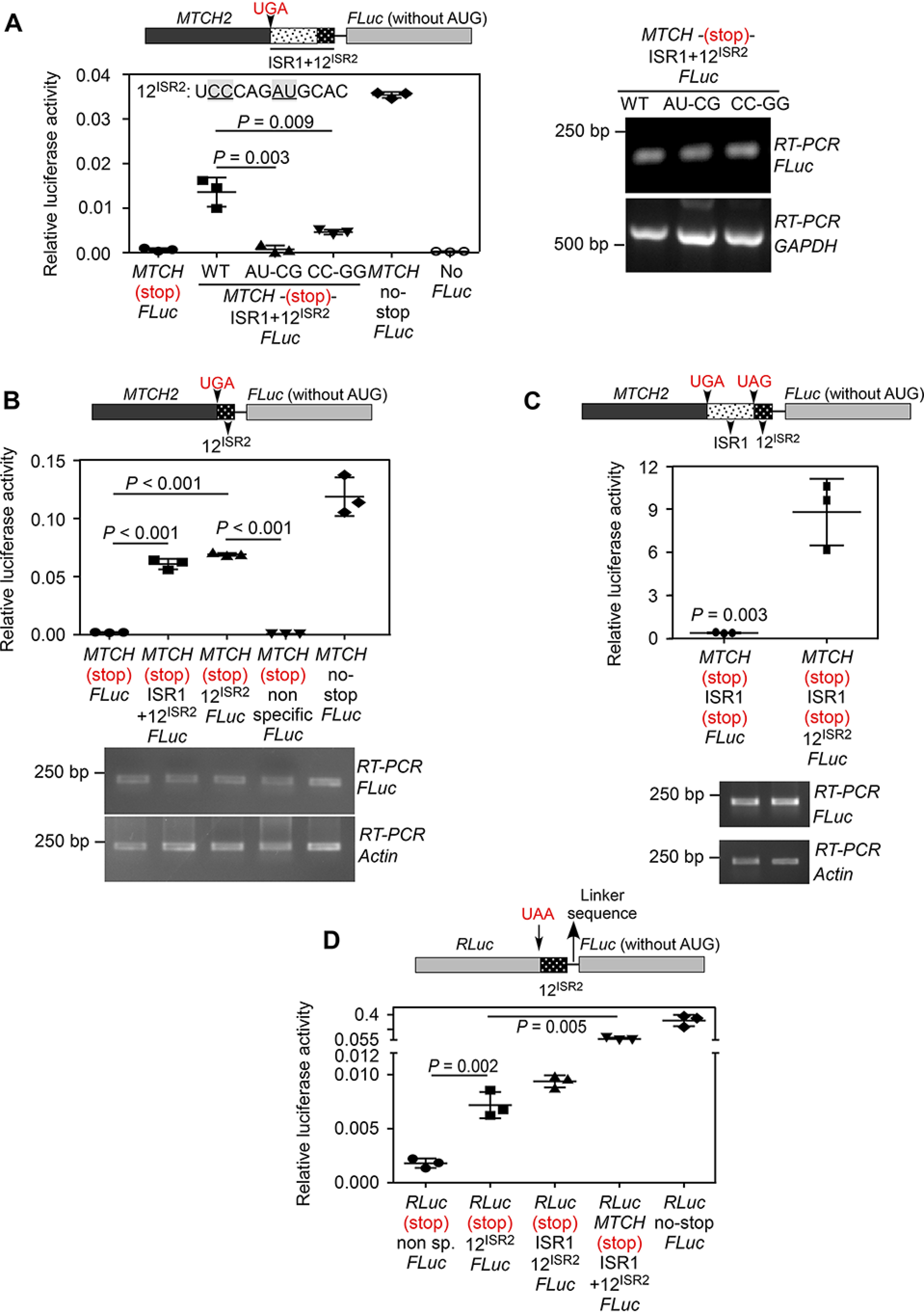
First 12 nucleotides of ISR2 (12ISR2) constitutes the RNA signal necessary for the SCR of MTCH2. A, mutations in 12ISR2 reduce read-through across the canonical stop codon. Constructs having ISR1 and mutated 12ISR2 were transfected in HEK293 cells and luciferase-based read-through assay was performed. Sequence of 12ISR2 is shown. Mutated residues are underlined. RT-PCR of luciferase mRNA is shown (right). B, 12ISR2 can drive read-through across the canonical stop codon of MTCH2. A construct having just 12ISR2 between MTCH2 and FLuc was used (schematic). Results of luciferase-based read-through assay carried out in HEK293 cells are shown. C, 12ISR2 can drive double-SCR of MTCH2. Construct having ISR1 + 12ISR2 along with the two stop codons between MTCH2 and FLuc was used (schematic). Results of luciferase-based read-through assay carried out in HEK293 cells are shown. D, 12ISR2 is sufficient to drive SCR in a heterologous context. 12ISR2 sequence was cloned between RLuc and FLuc such that all were in-frame. FLuc activity is expected only if the 12ISR2 sequence drives read-through across the stop codon of RLuc. Results of luciferase-based read-through assay carried out in HEK293 cells are shown. All graphs show mean ± S.D. (n = 3). Results are representative of at least three independent experiments done in triplicate. Two-tailed Student's t test was used to calculate the p value. MTCH, MTCH2; 12ISR2, first 12 nucleotides of ISR2 (UCCCAGAUGCAC).
