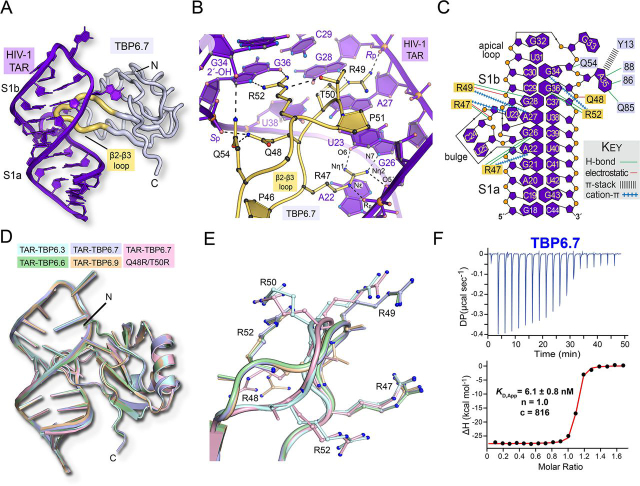
Figure 2.
Structural overview of lab-evolved protein TBP6.7, superpositions of HIV TAR-TBP co-crystal structures of this investigation, and TAR binding to TBP6.7.A, ribbon diagram depicting the overall fold of the TAR-TBP6.7 complex reveals entry of the β2-β3 loop into the major groove (PDB entry 6CMN) (28). B, close-up view of the lab-evolved β2-β3 loop of TBP6.7 showing TAR readout that includes Arg-47, which reads the Hoogsteen edge of Gua26 and the Uri23 backbone; Arg-49, which recognizes N7 of Gua28 and the phosphate backbone; and Arg-52, which reads the Hoogsteen edge of Gua36. Here and elsewhere, putative interactions are shown by dashed lines. C, overview of chemical interactions between the TAR-TBP6.7 complex that are representative of the various RNA-protein interactions of other TBPs in the current investigation. D, pairwise all-atom superposition of the co-crystal structures of this investigation upon the TAR-TBP6.7 complex. The average r.m.s.d. was 0.43 Å. The overall three-dimensional fold of each TBP is similar to TBP6.7. TAR RNA also reveals structurally similar details (Fig. S2). E, close-up view of the superimposed β2-β3 loops from D. Arginine placement affects the loop conformation and the mode of TAR recognition. The average loop r.m.s.d. was 0.78 Å. F, representative ITC thermogram of TBP6.7 titrated into TAR. The apparent equilibrium dissociation constant (KD) is shown, along with the stoichiometry (n), and the c value to indicate the quality of the binding model fit (34). Here and elsewhere, the representative single-run ITC parameters shown on thermograms differ from Table 2, which reports average values from duplicate titrations.