Figure 3.
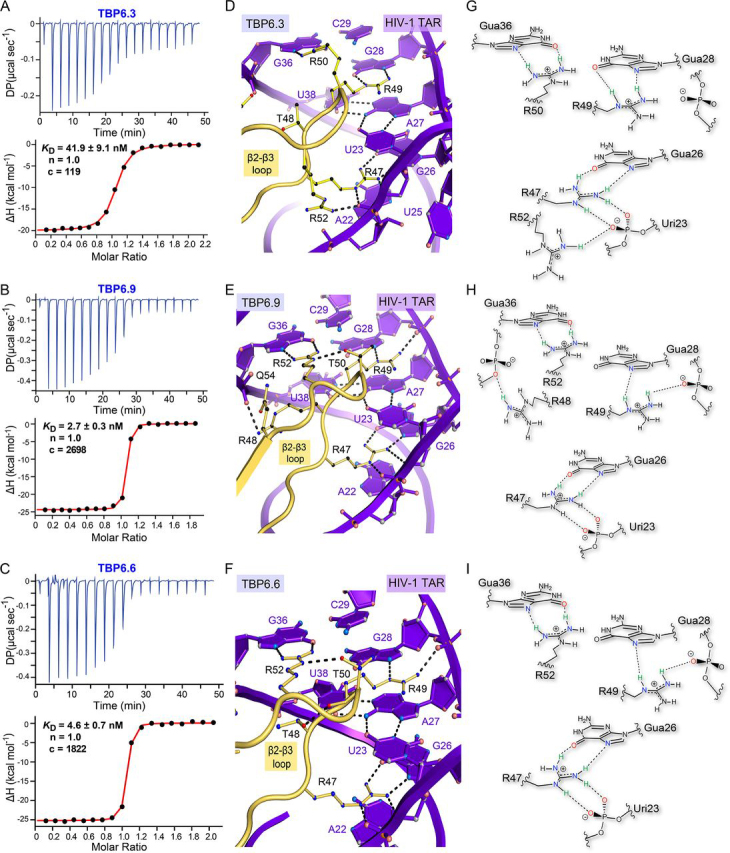
Affinity analysis of TBP variants for HIV-1 TAR and close-up views of TAR-TBP co-crystal structures revealing arginine-mediated RNA recognition.A, representative ITC thermogram of TBP6.3 titrated into TAR. B, representative ITC thermogram of TBP6.9 titrated into TAR. C, representative ITC thermogram of TBP6.6 titrated into TAR. D, close-up view of the lab-evolved β2-β3 loop of TBP6.3 showing TAR readout by four arginines. Arg-47 and Arg-49 retain interactions similar to TBP6.7 (Fig. 2, B and C), but Arg-52 is displaced by Arg-50 to recognize Gua36. As a result, Arg-52 now binds the backbone pro-Rp oxygen of Uri23. E, close-up view of the lab-evolved β2-β3 loop of TBP6.9 showing TAR readout by four arginines, including three consecutive arginines. Arg-48 interacts with the backbone at the pro-Sp oxygen of Gua36, similar to Gln-48 of TBP6.7, whereas Arg-47, Arg-49, and Arg-52 retain Hoogsteen-edge readout similar to TBP6.7 (Fig. 2, B and C). F, close-up view of the lab-evolved β2-β3 loop of TBP6.6 showing TAR readout by three arginines. The mode of interaction is comparable with TBP6.7 (Fig. 2B), except that Thr-48 and Thr-50 engage in stabilizing side chain-to-backbone interactions. G, schematic diagram depicting arginine interactions between TBP6.3 and TAR in D. H, schematic diagram depicting arginine interactions between TBP6.9 and TAR in E. I, schematic diagram depicting arginine interactions between TBP6.6 and TAR in F.
