Figure 4.
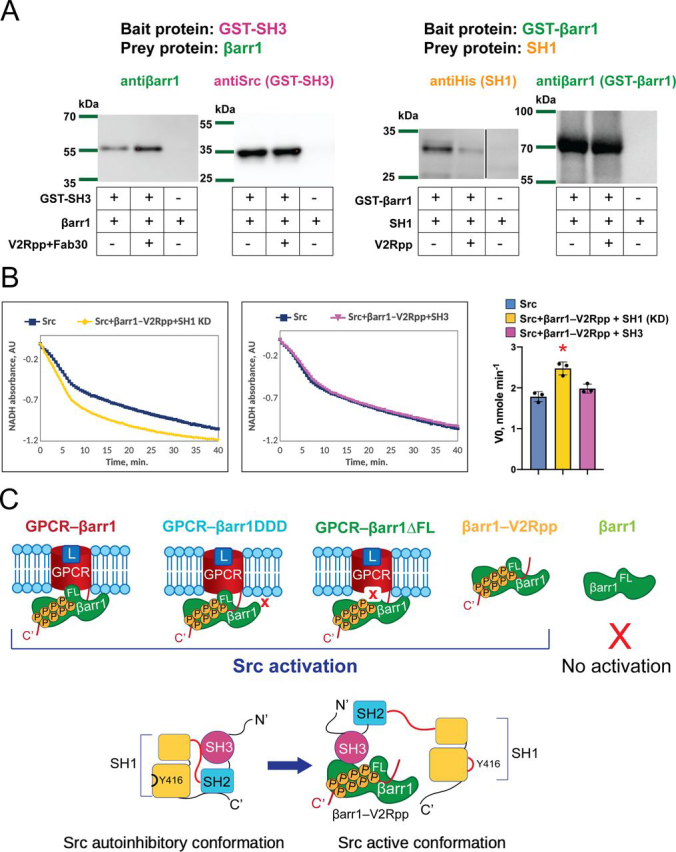
GPCR-activated βarr1 activates Src by interacting with its SH3 domain.A, βarr1 interacts with SH3 and SH1 domains of Src. Left panel, GST-pulldown assay of SH3 domain and βarr1. Right panel, GST-pulldown assay of SH1 domain and βarr1. The right lane separated by a black line was spliced from a non-neighboring lane of the same blot. Data shown are representative of three independent experiments. B, addition of an excess of SH3 domain blocks βarr1-mediated activation of Src. Left and middle panels show representative progress curves of NADH oxidation coupled to peptide phosphorylation by Src in the presence of βarr1–V2Rpp and excess SH1 kinase dead (SH1 KD) (left panel) or SH3 domains (middle panel) measured by continuous kinase colorimetric assay. Right panel, initial velocity of peptide phosphorylation by Src in the presence of βarr1–V2Rpp and excess of SH3 or SH1 KD domains. Individual data show mean ± S.D. of three independent experiments. Statistical differences were determined by Kruskal-Wallis test (one-way ANOVA on ranks) and Dunn's multiple comparison test (*, p < 0.05 as compared with Src alone). Optimal Src peptide (AEEEIYGEFEAKKKK) is used at a concentration of 250 μm, Src is used at a concentration of 200 nm, SH3 and SH1 KD are used at a concentration of 6 μm. βarr1–V2Rpp (stabilized by Fab30) is used at a concentration of 1.2 μm. C, conformational basis of βarr1-mediated activation of Src. Various tail-bound βarr1 conformations interact with SH3 domain of Src and disrupt the autoinhibited conformation of the enzyme (L, ligand; FL, finger loop).
