Abstract
Mechanical signals play a central role in cell fate determination and differentiation in both physiologic and pathologic circumstances. Such signals may be delivered using materials to generate discrete microenvironments for the purposes of tissue regeneration and have garnered increasing attention in recent years. Unlike the addition of progenitor cells or growth factors, delivery of a microenvironment is particularly attractive in that it may reduce the known untoward consequences of the former two strategies, such as excessive proliferation and potential malignant transformation. Additionally, the ability to spatially modulate fabrication of materials allows for the creation of multiple microenvironments, particularly attractive for regenerating complex tissues. While many regenerative materials have been developed and tested for augmentation of specific cellular responses, the intersection between cell biology and material interactions have been difficult to dissect due to the complexity of both physical and chemical interactions. Specifically, modulating materials to target individual signaling pathways is an avenue of inter-disciplinary research that may lead to a more effective method of optimizing regenerative materials. In this work, we aim to summarize the major mechanotransduction pathways for osteogenic differentiation and to consolidate the known materials and material properties that activate such pathways.
Keywords: materials, mechanotransduction, osteogenesis, osteogenic differentiation
Introduction
One of the most exciting developments in regenerative technology is the increasing understanding that mechanical signals are essential, physiologic mechanisms for the elaboration of various growth and differentiation factors, directing progenitor cell differentiation and the development of specific tissue identities. This development has sparked significant interest in the incorporation of materials into regenerative strategies and challenged the classical paradigm for regenerative research.
Despite 30 years of research, the paradigm of progenitor cell and growth factor delivery on materials has not been realized in the realm of surgical and clinical practicality. In skeletal reconstruction, the idea of harvesting stem cells for ex vivo expansion and growth is highly impractical in practice as well as in expense because autologous methods for bone grafting or transfer of free-vascularized bone are available, despite their morbidity. While the idea of growth factor delivery for augmentation of skeletal regeneration has been of great interest, the two United States Food and Drug Administration (FDA)-approved growth factors for bone have shown that physiologic dysregulation with supraphysiologic dosages of single factors is not likely to be the ideal strategy for regeneration. One example is the fact that a decreasing usage of bone morphogenetic protein-2 (BMP-2) in anterior spinal fusion has occurred due to the reported increases in complications compared to traditional, growth factor-free, autologous bone grafting [1]. Some progress has been made in the incorporation, or doping, of bioactive small molecules into bone graft materials[2]. However, this approach has a number of important limitations – the decline in the biological activity of the drug, potential damage to adjacent structures, and burst release all remain significant challenges that limit the clinical application of this approach. In combination, a strategy that can eliminate ex vivo progenitor cell expansion and growth factor administration would significantly reduce cost, potentially reduce the morbidity of additional procedures, reduce complications from uncontrolled cellular responses, and increase practicality in surgery.
The induction of specific cellular responses to materials has recently become a robust and growing area of research. Work by Dupont et al. and others has elucidated a number of the requisite signaling mechanisms for detection of microenvironmental changes in mechanical properties such as stiffness and translation of such signals intracellularly to effect cell biological changes including osteogenic differentiation, fibrosis, and metastatic potential of cancer cells [3–8]. For the purposes of regenerative material design, one avenue for optimization of materials may be modulating material properties to target mechanotransduction signaling pathways known to promote specific cellular processes. In this work, we review the relevant mechanotransduction pathways for osteogenic differentiation and consolidate the reported cell biological changes in materials investigated for bone regeneration.
Focal Adhesions, the Cytoskeleton, and Detecting the Extracellular Environment
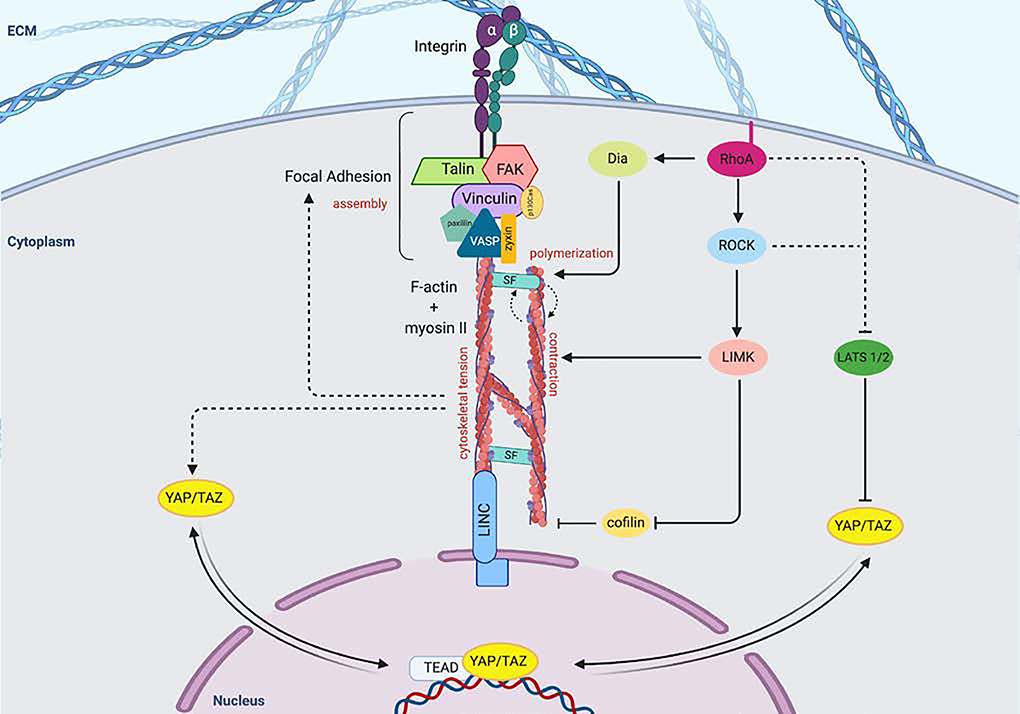
Cellular detection of biomechanical properties frequently begins at focal adhesions (FAs) [5]. One of the key members of FAs is the integrin αβ heterodimer at the cell surface [9]. The extracellular domains of integrins interact with the extracellular matrix (ECM) and adjacent cells, and their cytoplasmic domains are involved in the assembly of signaling complexes with intracellular cytoskeletal proteins [10] (Figure 1). The cytoplasmic tails of integrins are connected with cytoplasmic F-actin bundles via a number of docking proteins, including talin and vinculin. Force loading at the FA causes a conformational change in talin proteins, revealing additional binding domains for vinculin [11]. Vinculin binding facilitates the localization of additional integrins, and the presence of the vinculin tail domain is necessary to propagate force from the FA to the actin cytoskeleton [12]. Other proteins like zyxin and actinin also act cooperatively to stabilize actin polymerization and cross-linking, though other proteins including p130Cas and paxillin are also important components of FA assembly [13].
Figure 1.
Extracellular mechanical signals lead to focal adhesion assembly and activation of intracellular signaling pathways.
Focal adhesions (FA) consist of integrin clusters, which undergo morphological changes that recruit FAK and docking proteins including talin, vinculin, paxillin, as well as p130Ca adaptor proteins that assist in transferring mechanical signals to the actin cytoskeleton. The FAs inner core consists of VASP, zyxin and actinin, which regulate actin assembly. Force generation occurs by F-actin sliding on myosin II, and stress fibers (SFs), which are comprised of ɑ-actinin, fascin and filamin, stabilize this interaction. ROCK regulates the formation and tension of actin bundles and SFs and also phosphorylates and activates LIM kinase (LIMK), which in turn phosphorylates and inhibits the actin-severing protein cofilin. RhoA activation also promotes actin assembly through binding Diaphenous (Dia), which directly promotes actin polymerization, as well as inhibition of LATS 1/2. Inhibition of LATS 1/2 allows YAP/TAZ to translocate to the nucleus in response to cytoskeletal tension and contractile force. Nuclear mechanotransduction also occurs via transmission through LINC complex. FAK, focal adhesion kinase; VASP, vasodilator-stimulated phosphoprotein; SF, stress fiber; Dia, diaphanous; ROCK, Rho-associated protein kinase; LIMK, LIM kinase; LATS, Large tumor suppressor; Linker of Nucleoskeleton and Cytoskeleton (LINC) complex; TEAD, TEA domain family member.
Focal adhesion kinase (FAK) also plays an important and early role in determining the actin-based response to external mechanical stimuli. FAK contains a focal-adhesion-targeting (FAT) sequence that engages with talin and paxillin. In response to tension at the FA, FAK is recruited and autophosphorylated. Autophosphorylation of FAK further activates cytoskeletal contraction (as well as other intracellular mechanotransducing proteins), and both cytoskeletal contraction and spreading reinforce FAK activation [14]. Thus, the cellular response to mechanical stress occurs in a feed-forward loop – the formation of FAs promotes the growth of actin bundles, and cytoskeletal contractility stabilizes and facilitates maturation of FAs [15, 16]. The mechanism of cytoskeletal contractility occurs via F-actin sliding on myosin II, whilst being held together by stress fibers (SFs), which are comprised of α-actinin, fascin, and filamin. Stress fibers are responsible for propagating force from the ECM into the cell by pulling on FAs, and SFs and FAs cooperatively stabilize one another [17]. However, the primary means by which the actin cytoskeleton is stabilized during the application of tensile force is via inhibition of cofilin, which normally acts to sever F-actin fibers. Cofilin is inactivated during mechanical stimulation via phosphorylation by LIM kinase (LIMK), which is a kinase that is activated by the small GTPase RhoA via Rho-associated kinase (ROCK) [18]. RhoA also regulates the formation and tension of actin bundles and their associated SFs via activation of the formin Diaphanous (Dia). RhoA binds to the GTPase binding domain of Dia, facilitating a conformational change to release autoinhibitory and autoregulatory domains. ROCK also phosphorylates myosin light chain (MLC), which activates myosin II ATPase and generates contraction [19]. Cells thereby respond to the rigidity of the ECM substratum by adjusting the tension within the cytoplasm and organization of SFs such that cell spreading is accompanied by increased pulling forces against the ECM [20, 21]. Inhibitor studies have demonstrated that the organization and reorganization of actin polymerization and SF formation are essential to the activation of mechanotransduction pathways and the resultant cellular processes such as proliferation and differentiation [15, 16].
YAP/TAZ: Master Transcriptional Integrators of Mechanical Signals
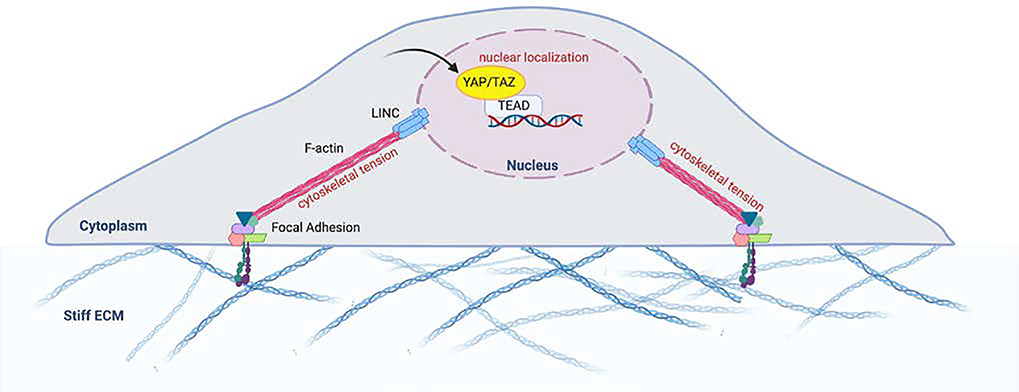
Though many of the mechanisms translating mechanical stimuli into biochemical stimuli are subjects of an ongoing investigation, Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ), traditional downstream effectors of the Hippo signaling pathway, have been demonstrated to play integral roles in delivering the signal of mechanical changes in the cellular microenvironment to within the nucleus [3, 22]. However, Hippo-independent pathways for YAP/TAZ activity have garnered the most attention recently with respect to cell fate determination secondary to mechanical cues [3, 23]. Generally, YAP/TAZ translocate from the cytoplasm to the nucleus when cells are plated on rigid ECM and/or following the application of mechanical force [24]. Elosegui-Artola et al. demonstrated that this partially occurs via mechanical forces being delivered via direct contact between the F-actin cytoskeleton and the nucleus via the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex. Physical deformation via the cytoskeleton leads to the opening of nuclear pores as well as an increase in active YAP import (Figure 2) [25]. YAP/TAZ thereby shuttle between the nucleus and the cytoplasm in response to stimuli from the ECM via regulation of the actomyosin cytoskeleton. Overall, localization of YAP/TAZ within the cytoplasm is indicative of inhibition. Upon nuclear translocation, YAP/TAZ interact with stage- and cell-specific Transcriptional enhanced associate domain (TEAD) transcription factors to alter the expression of genes like ankyrin repeat domain 1 (ANKRD1) and connective tissue growth factor (CTGF)[26]. YAP/TAZ have been found to be key regulators of cell proliferation, survival, and differentiation, and a growing body of literature has indicated their role in several key signaling pathways, including Hippo, Wnt, and BMP [3, 6, 7, 15]. Using Rho inhibitor C3 and latrunculin A, Dupont et al. showed that nuclear YAP/TAZ required Rho and the actin cytoskeleton for maintenance, which was independent of the Hippo/LATS cascade [3]. Also, inhibition of ROCK using Y27632 similarly demonstrated that the cytoskeletal tension via F-actin contractility is required for YAP/TAZ nuclear localization [6].
Figure 2.
Deformation of the cytoskeleton and tension leads to passive and active YAP import Extracellular signals are also transmitted via deformational changes in the cytoskeleton. Direct contact between the F-actin cytoskeleton and the nucleus via the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex allows direct propagation of mechanical force that results in the opening of nuclear pores, as well as an increase in active YAP import. Adapted from: Control of cellular responses to mechanical cues through YAP/TAZ regulation. Dasgupta et al. 2019.
Hippo-dependent pathways have also been described for the activation of YAP and TAZ in response to mechanical signals. Central to Hippo signaling is the activation of large tumor suppressor gene 1 and 2 (LATS1/2) which then control YAP/TAZ activation and nuclear translocation. Using mammary epithelial cell lines, Kim and Gumbiner demonstrated that adhesion to fibronectin and cell spreading resulted in the activation of YAP via FAK, Src, PI3K (phosphatidylinositol 4,5-bisphosphate 3-kinase), and PDK1 (3-phosphoinositide–dependent protein kinase 1) [27]. Similarly, other investigators have shown that durotaxis, cell movement based on stiffness, in hepatic stellate cells was dependent on FAK-mediated YAP activation [28]. Sabra and colleagues demonstrated that β1 integrin-dependent cell adhesion to the ECM was essential for proliferation of osteoblasts and murine embryonic fibroblasts via YAP nuclear translocation. YAP activation was mediated by recruitment of LATS1/2 to the plasma membrane upon β1 integrin binding, activation of the small GTPase RAC1, the activity of P21 (RAC1)-activated kinase (PAK), and inhibition of merlin [29]. Thus, the combination of these data suggests that multiple signals for YAP/TAZ activation may be concurrently activated by mechanical signals. Cell type and matrix components may also play a role in dominance of the individual pathways.
In all of these mechanisms, YAP/TAZ are generally nuclear and active when cells are spread over a stiff ECM, and are cytoplasmic and inactive when cells are placed on a soft ECM [15]. The relative importance of the various structures involved in F-actin-mediated localization of YAP/TAZ is yet to be fully established. The activation of YAP/TAZ has also been demonstrated to strongly affect cell differentiation into adipocytes or osteocytes (and cell differentiation more broadly), potentially even superseding that of cytokine signaling [15]. Knockdown of YAP in MSCs has been shown to induce adipogenesis while suppressing osteogenic differentiation [6]. Osteogenic differentiation of mesenchymal stem cells (MSCs) involves the interaction of YAP/TAZ with multiple key signaling pathways, including Wnt and BMP/Small Mothers Against Decapentaplegic (Smad)[30].
Relationship Between Wnt Signaling and Mechanical Signals in Osteogenic Differentiation
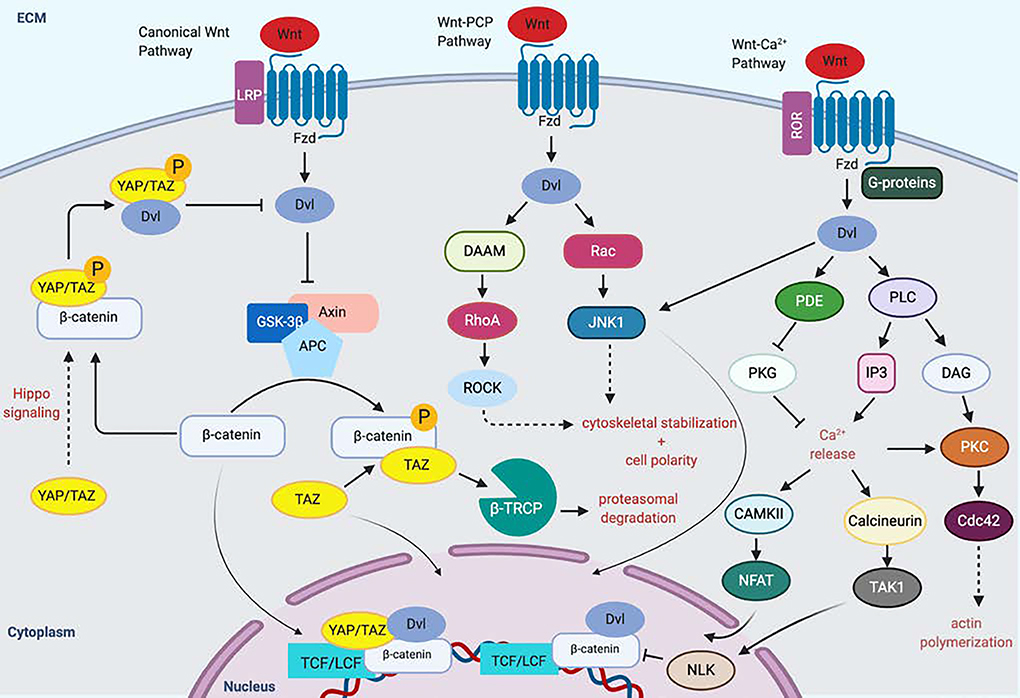
The Wnt pathway plays an important role in osteogenic differentiation initiated by mechanical stress. The canonical Wnt pathway involves binding of the Wnt ligand to the transmembrane receptor Frizzled (Fzd), which forms a complex with LDL Receptor Related Protein 5/6 (LRP5/6) [31]. The Wnt-Fzd binding activity causes Dishevelled (Dvl) to inhibit the function of the axin/adenomatous polyposis coli/glycogen synthase kinase-3β (Axin/APC/GSK-3β) complex, freeing β-catenin to translocate to the nucleus and act as a coactivator of transcription with the transcription factor/lymphoid enhancer-binding factor (TCF/LEF) family [32]. The role of the canonical Wnt pathway in osteogenesis has been demonstrated by extensive studies involving both inactivation or overexpression of β-catenin and LRP5 in humans and mice [33]. Overexpressing LRP5 promotes proliferation and osteogenesis in MSCs, while decreasing levels of Wnts and LRP5 inhibits Wnt pathway signaling [34]. Inactivation of β-catenin in osteoblasts causes osteopenia by affecting bone resorption rather than bone formation [35]. Work by Kang et al. demonstrated that transient activation of Wnt/β-catenin signaling in MSCs in vitro suppresses transcription of adipogenic transcription factor peroxisome proliferator-activated receptor-γ (PPAR-γ) and induces expression of other bone lineage genes such as Dlx5 and Osterix [36]. Aspects of both canonical and non-canonical Wnt signaling have been shown to play essential roles stimulating osteogenic differentiation of MSCs (and cell fate more generally), and they have numerous reciprocal interaction points with YAP/TAZ signaling [7, 31] (Figure 3).
Figure 3.
Wnt pathway activation and mechanotransduction promotes osteogenesis.
Binding of the Wnt ligand to the transmembrane receptor Frizzled (Fzd), forms a complex with LDL Receptor Related Protein 5/6 (LRP5/6), causing Dishevelled (Dvl) to inhibit the function of the Axin/Adenomatous polyposis coli/Glycogen Synthase Kinase-3β (Axin/APC/GSK3β). This frees β-catenin, which can then shuttle to the nucleus. Phosphorylated β-catenin serves as a binding scaffold for TAZ, enabling the degradation of the both TAZ and β-catening via the with β -TrCP/E3 ubiquitin-ligase degradation complex YAP/TAZ provide context-dependent upregulation or downregulation of the canonical Wnt pathway. The Wnt-planar cell polarity (PCP) pathway involves activation of Rho and Rac GTPases as well as ROCK and Jun N-terminal kinase (JNK) to modulate cytoskeletal organization and gene expression that alters cell polarity. This involves signaling through Rac, Rho small GTPases. The Wnt-Ca2+ pathway involves binding of Wnt to Fzd, causing G-protein mediated activation of phospholipase C (PLC). IP3 causes release of Ca2+ from the endoplasmic reticulum, which activate CAMKII and Calcineurin, as well as protein kinase C (PKC). These proteins d other downstream effectors like Cdc42, which promote actin polymerization, while NFAT and TAK1 boost the expression of several genes leading to a variety of functions, including osteogenesis. LRP, LDL Receptor Related Protein; Fzd, frizzled; Dvl, Dishevelled, GSK-3β, Glycogen Synthase Kinase-3β; TCF/LCF, transcription factor/lymphoid enhancer-binding factor; Axin/Adenomatous polyposis coli/Glycogen Synthase Kinase-3β (Axin/APC/GSK3β); β -TrCP, β-transducin repeats-containing proteins; DAAM, Disheveled-associated activator of morphogenesis; JNK c-Jun N-terminal kinase; PDE, phosphodiesterase; PKG; protein kinase G; PLC, phospholipase C; IP3, inositol 1,4,5- triphosphate; DAG; Diacylglycerol; PKC, protein kinase C; CAMKII Calcium/calmodulin-dependent protein kinase type II; NFAT, nuclear factor of activated T-cells; TAK1, Transforming growth factor β-activated kinase; Cdc42, cell division control protein 42 homolog
Unlike the canonical pathway, the non-canonical Wnt pathway is independent of β-catenin and signals through phospholipase C (PLC) and phosphokinase C (PKC), which regulate intracellular calcium release [37]. Wnt ligands like Wnt5a bind to receptor tyrosine kinase-like orphan receptor (Ror) 1/2 and activate β-catenin-independent signaling pathways such as the planar cell polarity (PCP) pathway and the calcium pathway. In the PCP pathway, Fzd is associated with Dvl and disheveled-associated activator of morphogenesis (Daam) to activate Rho, Rac, and Cdc42. In the calcium pathway, intracellular calcium increases through receptor-coupled G proteins and phospholipase C [38].
The discovery by Azzolin and colleagues that YAP/TAZ are components of the cytosolic β-catenin destruction complex was the key finding that connected mechanoregulation to the canonical Wnt pathway [39]. In the presence of canonical Wnt ligands such as Wnt3a, YAP/TAZ and β-catenin are dissociated from the destruction complex resulting in nuclear translocation and transactivation of downstream genes, including Runt-related transcription factor-2 (Runx2) [31, 40]. In contrast, the absence of Wnt sequesters YAP/TAZ in the cytosol on the destruction complex where YAP/TAZ is required for the recruitment of the E3 ubiquitin ligase β-transducin repeat-containing protein (β-TrCP), which then polyubiquitinates β-catenin, targeting it for proteasomal degradation.
Relationship Between BMP Signaling and Mechanical Signals in Osteogenic Differentiation
Signaling pathways downstream of transforming growth factor-β (TGF-β) superfamily of receptors, including the BMP receptors, are other major axes in osteogenic differentiation. Similar to the Wnt pathway, binding of the TGF-β/BMP receptors (TGF-βR/BMPR) to their cognate ligands may activate canonical or non-canonical intracellular signaling cascades. In the canonical pathway, the receptor Smads (Smad1/5/8 for BMPR and Smad2/3 for TGF-β) are phosphorylated and dissociate from the receptor to complex with the co-Smad, Smad4. This complex then translocates to the nucleus and activates transcription of target genes, including Runx2 (Figure 4). Negative regulation of receptor Smads occurs via binding of the inhibitory Smads (Smad6/7), which prevent co-Smad association and targets the receptor Smads for proteasomal degradation by complexing with the E3 ubiquitin ligase Smad ubiquitin regulatory factor-1 (Smurf1) [7, 41]. In the non-canonical pathways, activation of a myriad of intracellular pathways may occur including extracellular signal-regulated kinase-1/2 (ERK1/2), p38 mitogen activated protein kinase (MAPK), and Akt [42]. Such pathways have all been associated with osteogenic differentiation in a context dependent manner, however, the intersection between BMPR/TGF-βR signaling and mechanotransduction has been via the canonical BMPR pathway.
Figure 4.
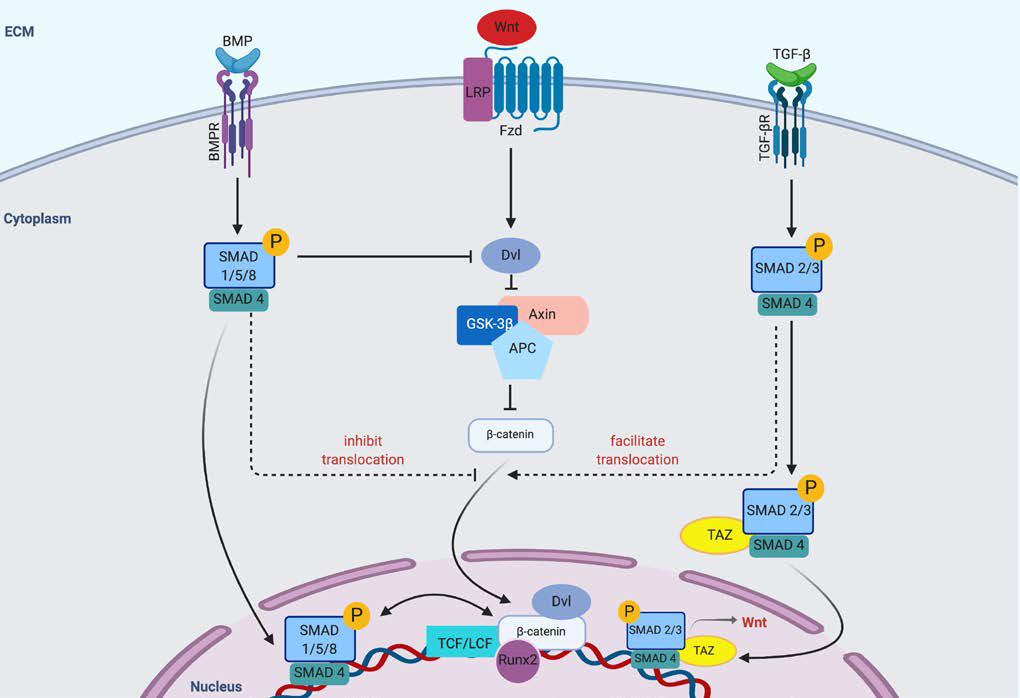
Crosstalk between the BMP/Smad, Wnt, and TGF-β pathways. Crosstalk occurs at multiple levels following ligand activation of the varying receptors. β-catenin translocation to the nucleus is affected by Smad complex activation or suppression by the TGF- β and BMP signaling pathways.
Both direct and indirect interactions between YAP/TAZ with receptor Smads within the BMPR pathway have been reported. Alarcon et. al demonstrated that YAP increases BMP signaling via association with Smad1/5, and that YAP co-precipitates with Smad5 on BMP target sites. In addition, YAP depletion is inhibited by the induction of target genes by BMP [43]. In TGF-β signaling, TAZ was shown to be required for nuclear accumulation and transcriptional activity of the receptor Smads (Smad2/3) [44]. These reports identified TAZ (but not YAP) as required for Smad2/3 nuclear accumulation and transcriptional activity in response to TGF-β signaling, as well as connectivity between the YAP/TAZ mechanotransduction pathway and BMP/Smad mediated osteogenesis [7].
Crosstalk between Wnt and BMP/Smad Signaling
Interaction between the Wnt pathway and the BMP/Smad pathways in osteoblastic differentiation occurs at multiple points and is an ongoing area of investigation. Using a variety of pluriportent mesenchymal cell lines, Rawadi and colleagues demonstrated that expression of ALP by Wnt3a was independent of BMPR signaling, however, the ALP expression induced by exogenous BMP-2 required the activity of Wnt/LRP5 downstream signals [45]. The effect of the canonical Wnt pathway on BMPR signaling has been found to be both synergistic and antagonistic. Axin2 knockout mice, which result in increased β-catenin activity, have been found to display a phenotype of increased bone formation with increases in BMP2 and BMP4 gene expression and Smad1/5 phosphorylation [46]. Mbalaviele et al. demonstrated that truncated β-catenin (in which the GSK-3 phosphorylation sites were deleted) did not enhance ALP activity on its own, but in the presence of BMP-2, resulted in synergistically increased ALP activity, OCN expression, matrix mineralization, and new bone formation when injected into mouse calvaria in vivo [47]. In primary calvarial osteoblasts, BMP-2 enhanced Wnt signaling via upregulation of several Wnt ligands and receptors [48]. Conversely, the expression of inhibitors of BMP signaling, BMP3 and Protein Related to DAN and Cerberus (PRDN), have both been found to be dependent upon β-catenin activity [49]. Taken together, the intersection between the canonical Wnt and BMPR signaling pathways are complex. Clearly, there is interdependency upon each other, however, negative regulatory mechanisms that are incompletely understood also exist.
Material-Induced Mechanotransduction and Osteogenic Differentiation
The introduction of materials into regenerative strategies has occurred several decades ago. Initially thought of as inert carriers, materials have gained significant attention as mediators of cell differentiation and proliferation via its intrinsic properties. The rationale of how properties affect cell behavior is now clearly related to the activation of specific mechanotransduction pathways, thus, suggesting that modulation of properties may augment or diminish the respective pathways (Table 1).
Table 1.
Materials and Their Effects
| Materials/Substrates | Factor | Properties | Cell Type | Effects | Reference |
|---|---|---|---|---|---|
| Inert PAAm gels with varying concentrations of bis-acrylamide | Stiffness | Elastic modulus: 0.1, 1, 11, and 34 kPa | MSCs | Increased stiffness promoted the expression of FA components. Addition of blebbistatin disrupted the actin cytoskeleton, suggesting that actin cytoskeleton formation is dependent on non-muscle myosin II | Engler et al., 2006 |
| PAAm with fibronectin coating | Stiffness | Intermediate (~8 kPa) and stiff (~100 kPa) polyacrylamide (PAA) gel substrates, and glass (~1 GPa) | NIH3T3 cells | Higher stiffness led to a decrease in key molecules in the cell membrane, including talin and vinculin | Stutchbury et al., 2017 |
| PAAm with collagen-I coating | Stiffness | Young’s modulus: 1.46 and 26.12 kPa | UC-MSCs | Expression of the integrin subunits α2 and β1 increased with higher stiffness | Witkowska-Zimny et al., 2014 |
| PDMS matrices | Stiffness | Substrate stiffness varied with “1:5, 1:15, 1:30, and 1:45 ratios [curing agent (Sylgard 184, Corning, NY, USA) vs oligomeric base]” | hSCAPs | Cells on stiffer substrates showed increased levels of fibronectin, which interacted with FAK and paxillin, promoting nuclear accumulation of β-catenin and suggesting the involvement of the Wnt pathway | Zhou et al., 2019 |
| PDMS substrates | Stiffness | 6, 16, 54, 54, and 135 kPa | DPSCs | Higher stiffness enhanced the expression of β-catenin and decreased the expression of GSK-3β, indicating the involvement of the Wnt pathway | Liu et al., 2019 |
| PEG hydrogels functionalized with RGD | Stiffness | Elastic modulus: 13.67 and 423.89 kPa | Pre-osteoblastic MC3T3-E1 cells | Increased stiffness enhanced the expression of ALP, OCN, and BSP, and the activity of p44/42 MAPK (19). Stiffer matrices also promoted FAK activation, increasing RhoA activity and cell contractility via ROCK (20) | Khatiwala et al., 2007 and Khatiwala et al., 2009 |
| Collagen-coated hydrophobic PDMS and hydrophilic PEO-PDMS | Stiffness | Elastic modulus: less than 1 to over 2000 kPA | hMSCs | Ligand assembly driven by surface energy promoted osteogenic differentiation on soft, hydrophobic substrates, even though the contractility of cells decreased | Razafiarison et al., 2018 |
| Inorganic substrate (Ti and HAP) with collagen | Stiffness | Substrates: Pure collagen, Ti with collagen, and HAP with collagen | MSCs | Pure collagen substrate inhibited osteogenic differentiation, but pristine Ti (greatest stiffness) induced the highest expression of myosin II. HAP led to increased collagen self-assembly and collagen fibrous network formation, compared to pristine Ti. HAP-collagen led to the highest promotion of osteogenic differentiation via canonical Wnt/β-catenin pathway | He et al., 2016 |
| PEG hydrogels | Stiffness | Scaffold modulus: 0.2 to 59 kPa | hBMSCs | Osteogenic differentiation was promoted on stiffer substrates. Addition of Arginie-Glycine-Aspartate-Serine peptide inhibited osteogenesis by blocking integrin receptors, suggesting that integrins are necessary for osteogenesis on these substrates | Parekh et al., 2011 |
| RGD-modified 3D hydrogels | Stiffness | Elastic modulus: 2.5 to 110 kPa | mMSCs | Inhibiting myosin contractile machinery diminished stiffness-dependent bond formation. Decreased bond formation reduced osteogenic differentiation | Huebsch et al., 2010 |
| 3D scaffold consisting of PLA and HAP | Stiffness | PLA scaffolds--mean porosity: 64.2%, pore sizes: 224 μm, SA-to-volume ratio: 35.8 mm−1, thickness: 2.4 mm; PLA/HAP scaffolds--mena porosity: 65.2%, pore sizes: 249 μm, SA-to-volume ratio: 34.8 mm−1, thickness: 2.4 mm | hMSCs | Increased stiffness promoted mineralization, ALP activity, and osteoblastic differentiation | Persson et al., 2018 |
| 2D monolayers, 3D scaffolds | Geometry | 2D monolayer: tissue culture polystyrene (TCPS); 3D scaffolds: fabricated from PEOT/PBT by utilizing fused deposition modeling (FDM) or electrospinning (ES) | MSCs | Cells on 3D scaffolds with specific soluble factors showed similar differentiation to those on 2D monolayers | Leferink et al., 2015 |
| Poly(trimethylene carbonate)-based 3D microtopographic culture chips with concave and convex spherical structures | Geometry | Principal curvatures of convex or concave 3D structures: κ = 1/125, 1/175, 1/250, and 1/375 μm−1 | MSCs | Pulling and pushing forces via cytoskeleton tension promoted osteogenic differentiation on concave 3D structures | Werner et al., 2017 |
| Fibronectin-coated PDMS substrates | Geometry | Sizes of islands: 1024, 2025, or 10,000 μm2 | MSCs | Larger islands promoted osteoblastic differentiation with increased cell spreading, RhoA and ROCK signaling, and actin-myosin-generated tension | McBeath et al., 2004 |
| Octadecanethiolate on a glass coverslip coated with gold, modified with a tri-(ethylene glycol)-terminated monolayer followed by fibronectin | Geometry | Areas of mixed shapes: 1000, 2500, 5000 μm2 | MSCs | Cells on larger shapes showed increased cell spreading and osteogenic differentiation via actinomyosin contractility, JNK, ERK1/2, and Wnt signaling | Kilian et al., 2010 |
| Biodegradable patch with micro-scale grooves | Geometry | Patch pattern: parallel direction (0°) and perpendicular direction (90°) | Osteogenic progenitor cells | Patch patterned in perpendicular direction promoted cell migration, compared to patch patterned in parallel direction | Lee, M. S., 2018 |
| Poly-dopamine coated PLLA nanofibers | Geometry | Nanofiber orientation: parallel direction (0°), perpendicular direction (90°), and randomly-oriented fibers | MSCs | Nanofibers in parallel direction showed faster cell migration. Aligned fibers showed enhanced bone regeneration in nanofiber direction, compared to randomly-oriented fibers | Lee et al., 2014 |
| Surface coated with 1D Mo3Se3- single chain atomic crystals | Geometry | MC3T3-E1 osteoblasts | Mo3Se3- coating promoted an increase in osteoblastic proliferation (396.2 ± 8.1%) | Dalby et al., 2014 | |
| AuNPs | Geometry | Diameters of nanospheres, nanostars, and nanorods: 40, 70, and 110 nm | MSCs | Particular AuNP shapes and sizes promoted osteogenic differentiation, mediated by YAP. Specifically, sphere-40, sphere-70, and rod-70 enhanced ALP activity, calcium deposition, and osteogenic gene expression, while rod-40 decreased them | Li et al., 2016 |
| Hierarchical macropore/nanopore Ti substrate | Geometry | PT surface (control): no micropores or nanopores could be seen; MT surface: macroporous but no apparent nano structures; MNT surface: both macropores and nanowires present | Bone marrow-derived MSCs (BMSCs) | Hierarchical MNT substrate increased FA development, cytoskeletal tension, YAP activity, and YAP nuclear translocation, compared to control substrate | Pan et al., 2017 |
| HSTC | Geometry | Average nanotube diameter: 30 nm, approximate nanotube wall thickness: 10 nm | MSCs | Hierarchical structure promoted cell adhesion, OPN and OCN expression, and osteogenic differentiation, compared to control structure | Xie et al., 2014 |
| Hierarchical MNRs | Geometry | Interrod spacing size: 33 to 300 nm | MSCs | Interrod spacing size of larger than 137 nm demonstrated reduced cell adhesion and proliferation, suppressing osteogenic differentiation in vitro and in vivo | Zhou et al., 2016 |
| Hierarchical MNRs | Geometry | Nanorod spacings: approx. 30, 70, and 150 nm | MSCs | Substrate with 70 nm nanorod diameter showed increased Wnt3a and LRP6 expression and decreased Dkk1, Dkk2, sFRP2, and sFRP1 expression, suggesting that it promoted osteogenic differentiation via Wnt/β-catenin pathway | Zhou et al., 2018 |
| Polymerized collagen gel with a protein fibrillar microarchitecture | Geometry/Stiffness | Bulk stiffness: 16.4 to 151.5 Pa, local fiber stiffness: 1.1 to 9.3 kPa | MSCs | Gels with shorter fiber lengths and higher fiber stiffness suppressed the transmission of traction forces and formation of FAs, inhibiting cell migration, spreading, and proliferation. They tended to promote adipogenic differentiation | Xie et al., 2017 |
| 3D macro-porous structure comprised of ELP | Geometry | Young’s modulus: 0.5 to 50.3 kPa | MSCs | Higher stiffness promoted both osteogenesis and adipogenesis, which were dependent on cellular orientation and substrate interactions | Haugh et al., 2018 |
| PAAm | Ligand Functionalization | Stiffness: 0.7, 10, 30, and 80 kPa; Ligands: collagen I, collagen IV, fibronectin, laminin | MSCs | Highest upregulation of Runx2 occurred in PAAm gels with collagen I | Rowlands et al., 2008 |
| Fibronectin-coated PAAm hydrogels | Ligand Functionalization | Fibronectin densities: low, intermediate, high; Stiffness: 3 and 38 kPa | hMSCs | Substrate stiffness altered YAP translocation only when ligands were patterned at intermediate densities. Cell spreading, F-actin formation, and osteogenic differentiation increased with higher stiffness | Stanton et al., 2019 |
| PS–PEO block copolymers with azide terminal ligand nanodomains | Ligand Functionalization | Nanodomain lateral spacings: 30 to 60 nm | MSCs | Smaller lateral spacings stimulated FAK and Src activation, leading to higher quanities of FAs and expression of osteogenic markers like Rac1, levels of β-catenin in cytoplasm, and nuclear traslocation of Runx2 and YAP/TAZ | Han et al., 2019 |
| Titanium | Ligand Functionalization | Coating: Uncoated, Allylamine plasma polymer (PPAAm), Collagen I | MG-63 osteoblastic cells | Both of the functionalized substrates increased the formation of FA and development of actin cytoskeleton | Finke et al., 2007 |
| Au surface modified by monolayers of alkanethiols | Ligand Functionalization | Functional groups: CH3, OH, COOH, NH2 | MC3T3-E1 cells | OH and NH2-terminated surfaces led to increased osteogenesis via β1-integrin-binding | Keselowsky et al., 2005 |
| Graphene coated on polyethylene terephthalate | Mixed and Dynamic Effects | Substrates: graphene, SiO2 | MSCs | Compared to cells on SiO2 substrate, those on graphene showed increased cell proliferation and osteogenic differentiation | Nayak et al., 2011 |
| PDMS substrates coated with a graphene monolayer | Mixed and Dynamic Effects | Substrates: PDMS only, PDMS with graphene monolayer | MSCs | MSCs on graphene scaffolds showed increased expression of FAK-p397, integrin, SMAD p1/5, Runx2, and OPN, indicating that the integrin/FAK pathway induced osteogenic differentiation | Xie et al., 2019 |
| 3D graphene foams | Mixed and Dynamic Effects | Substrate: 3D graphene foams; Control: tissue culture polystyrene (TCPS) | MSCs | Osteogenic differentiation was promoted on 3D graphene foams, with higher expression of the ostegenic markers OPN and OCN | Crowder et al., 2013 |
| 3D hydrogel matrix substrate | Mixed and Dynamic Effects | Chirality: left-handed and right-handed | MSCs | ECM with left-handed chiality enhanced the clustering of Itgα5, promoting contractility, FAK and ERK 1/2 cascades, and nuclear translocation of YAP | Wei et al., 2019 |
| Conducting polymer Ppy | Mixed and Dynamic Effects | Dynamic switching between nanotips and nanotubes | MSCs | Dynamic switching between nanotips and nanotubes altered surface adhesion and increased osteogenic differentiation, independent of chemical signals and surface stiffness. It also promoted cytoskeleton organization and osteogenic gene expression | Wei et al., 2017 |
| 3D GelMA hydrogels | External mechanical stimulation | GelMA hydrogel concentrations: 5, 7.5, and 10%; Dynamic compression strain: 0, 10, 27, and 42% | hMSCs | 5% GelMA hydrogels promoted cell sensitivity to compressive strain, with the cells under 42% strain showing the highest increase in osteogenesis and enhanced expression of osteogenic proteins including Runx2 and OPN | Seo et al., 2018 |
| O-IOPs-MC | External mechanical stimulation | IOP-to-collagen mass ratio: 0, 0.25, 0.5, 0.67, and 1; Static Magnetic Field stimulation: 100 mT | MC3T3-E1 cells | Application of static magnetic field promoted ALP activities. These cells also displayed enhanced cell spreading and expression of osteogenic genes, suggesting that osteogenic differentiation was promoted via RhoA-ROCK pathways mediated by integrins | Zhuang et al., 2018 |
| 3D collagen glycoaminoglycan scaffolds | External mechanical stimulation | Stimulation via flow-perfusion bioreactor: 1 mL/minute for an hour, then 0.05 mL/minute for 7 hours | rMSCs | Mechanical stimulation led to dose-dependent increase in placental growth factor (PGF) expression via actin polymerization. Lower concentrations of PGF promoted osteogenesis while higher concentrations promoted osteoclastogenesis | McCoy et al., 2013 |
Stiffness
Synthetic hydrogels have been used extensively to determine the effects of substrate stiffness and dimensionality on cell behavior. This is partially due to the relative ease of tuning the physical and mechanical properties of these substances by altering their preparation conditions, thickness, coating and/or precursors, as compared to natural materials [50, 51]. Some of the most frequently used synthetic hydrogels used include polyacrylamide (PAAm), polyethylene glycol (PEG), and polydimethylsiloxane (PDMS). Engler et al. demonstrated that by altering the elastic modulus of PAAm (at 0.1, 1.0, 11, and 34 kilopascals (kPa)) via different concentrations of bis-acrylamide, one can control the differentiation of cells into specific lineages, a process that is at least partially dependent on non-muscle myosin II [20]. Greater substrate stiffness promoted osteogenic differentiation, as well as the expression of FA complex-associated proteins, including filamin, talin, and FAK. Addition of the myosin II inhibitor blebbistatin to cultures effectively inhibited this process by disrupting the actin cytoskeleton and intracellular tension. Other studies have demonstrated the impact of PAAm stiffness on molecules involved in mechanotransduction pathways linked to osteogenesis. By increasing the stiffness of PAAm via fibronectin coating, it was shown that the mobility of key molecules (e.g. talin, vinculin, FAK) at the membrane of NIH3T3 cells was significantly decreased [52]. In a separate study, mesenchymal stromal/stem cells isolated from Wharton’s jelly of the umbilical cord (UC-MSCs) were cultured on collagen I-coated PAAm with an elastic modulus of either 1.46 or 26.12 kPa [53]. The PAAm material with greater rigidity was shown to significantly increase osteogenic differentiation of UC-MSCs through α2 integrin-mediated mechanotransduction events. On PDMS matrices with variable stiffness, human apical papilla stem cells (hAPSCs) were found to secrete higher levels of fibronectin and be more prone to osteogenic differentiation on stiffer substrates [54]. Fibronectin engages with FAK and paxillin, which were both found to directly interact with β-catenin. Increased nuclear accumulation of β-catenin was also seen in hAPSCs cultured on stiffer PDMS, indicating the involvement of the Wnt pathway. In another study, dental pulp stem cells (DPSCs) were cultured on PDMS substrates of increasing stiffness (6, 16, 54, and 135 kPa). Greater stiffness of the PDMS substrate material was demonstrated to increase both cell proliferation and osteogenic differentiation. Increased expression of β-catenin and decreased expression of GSK-3β was observed, again indicating the involvement of the canonical Wnt pathway [55]. PEG hydrogels functionalized with RGD also can be used at different stiffness levels by varying amounts of the cross-linkable polymer Poly(ethylene glycol) diacrylate (PEGDA). In one study, functionalized PEG hydrogels with elastic moduli of 13.7 and 423.9 kPa were used to evaluate the impact of these properties on pre-osteoblastic MC3T3-E1 cells [56]. It was found that MC3T3-E1 cells cultured on the stiffer substrate expressed higher levels of ALP, osteocalcin (OCN), and bone sialoprotein (BSP), as well as greater p44/42 mitogen-activated protein kinase (MAPK) activity. In addition, inhibition of MAPK activity with the small molecular inhibitor PD98059 suppressed expression of OCN and BSP, indicating that MC3T3-E1 cells undergo osteogenic differentiation through a MAPK-dependent mechanism. Similarly, another study by the same authors using PEG hydrogels (also at 13.67 and 423.89 kPa) and MC3T3-E1 pre-osteoblasts evaluated the signaling pathways involved [57]. On stiffer ECM, MC3T3-E1 cells demonstrated increased osteogenesis as measured by Runx2 expression, ERK1/2 phosphorylation, OCN expression, ALP activity, and mineralized matrix deposition. Inhibition of the small GTPase RhoA, ROCK or MAPK blocked stiffness-induced osteogenic differentiation and mineralization. Based on these findings, the authors suggested that stiffer matrices increase FAK phosphorylation, which promotes RhoA activity and enhances contractility of cells via ROCK. Multiple other in vitro studies investigating the stiffness of substrate gel materials have yielded consistent data, demonstrating that materials with higher elastic moduli (and are in that dimension more similar to bone in vivo), generally promote osteogenic differentiation and osteogenesis [58, 59]. The recognition that varying material stiffness influences differential cell fate determination has been particularly attractive in the development of gradient materials for junctional tissues such as the osteochondral interface [60].
While most studies indicate that increased stiffness of PDMS promotes osteogenic differentiation, other factors like the surface energy of the biomaterial can, in some instances, alter MSC signaling, and ultimately, cell fate. Razafiarison et al. examined human MSCs (hMSCs) on collagen-coated hydrophobic PDMS and hydrophilic polyethylene-oxide-PDMS (PEO-PDMS) [61]. Even though cell contractility was diminished on soft versions of both substrates, surface energy-driven ligand assembly was able to facilitate osteogenic differentiation on hydrophobic, soft PDMS. These findings suggest that surface polarity can steer mechanotransduction and downstream cellular responses, potentially overriding signals generated by stiffness in certain circumstances.
Despite the multitude of evidence for a positive effect of stiff materials on osteogenic differentiation, one of the known challenges in material design for skeletal regeneration is the balance between stiffness and elasticity. While increasing stiffness confers osteogenic lineage differentiation, excessively stiff materials are brittle and prone to breakage upon compression. Several potential methods of balancing the requirement for two antagonistic requirements for bone regeneration while still promoting skeletal regeneration include alterations of other physical properties, such as patterning and topography, or chemical properties, such as inorganic content, via synthesis of composite materials.
Geometry, Patterning, and Topography
Many investigators have studied the influence of various geometries and micropatterning of material surfaces, including grooves, pits, pillars, and ridges, in order to evaluate their impact on cultured cells’ physiology and behavior. Werner et al. used stereolithography to develop poly(trimethylene carbonate)-based 3D microtopographic culture chips with concave and convex spherical structures [62]. Convex 3D structures induced osteogenic differentiation of MSCs through mechanotransduction. Cytoskeletal tension-mediated pulling or pushing force was generated in response to the concave or convex 3D material structure, which affected cell attachment to the surface and resulted in nuclear deformation. McBeath et al. studied the effects of cell spreading on fibronectin-coated islands of various sizes and determined that MSCs differentiated into osteoblasts when they were able to spread on larger islands and adipocytes when restricted to a round shape on smaller islands [58]. In another study, MSC differentiation was evaluated by patterning individual MSCs on a substrate material with 2D geometrical shapes to evaluate the effects on differentiation [63]. The material consisted of octadecanethiolate on a glass coverslip coated with gold, modified with a tri-(ethylene glycol)-terminated monolayer followed by fibronectin. Cells patterned on larger shapes underwent greater cell spreading and were more prone to osteogenic differentiation. Microarray and pathway inhibition studies determined that this process was dependent on actomyosin contractility, c-Jun N-terminal kinases 1/2 (JNK1/2), ERK1/2 and Wnt signaling. Other studies have examined the directionality of micropatterns and their effects on osteogenesis and osteoblast migration as it relates to bone regeneration. Lee et al. created a biodegradable patch with anisotropic micro-scale grooves that impacted the rate of osteoblast migration depending on the direction of the nanopattern [64]. Specifically, patches patterned in the perpendicular direction showed increased cell migration compared to patches patterned in the parallel direction. When implanted into mice that had a calvarial or tibial bone defect, this patch was able to accelerate bone formation and regeneration (with corresponding changes in the expression of ALP and osteopontin (OPN)), compared to flat patches without the pattern. On the other hand, a different group using poly-dopamine coated poly(l-lactic acid) (PLLA) nanofibers organized in a parallel direction found that this organization led to faster migration (10.46-fold) of MSCs compared to those in a perpendicular direction [65]. When aligned fibers were implanted into a calvarial defect mouse model, there was increased bone regeneration compared to randomly oriented fibers, with bone regenerating in the direction of the nanofibers. The discrepancy between the two studies suggests an underlying mechanistic difference in cells’ response to the anisotropic patterned material. Regardless, these studies indicate that the orientation of nanofibers can serve as a stimulation cue that guides cell migration in vitro and bone regeneration in vivo.
Physiological ECM is composed of a number of molecules that give it topography at a nanometer scale, which has also been shown to strongly influence cell behavior. Accordingly, many material substrates, including one-dimensional (1D) carbon nanotubes, gold nanowires, and silicon nanowires have been created to mimic the ECM’s nanotopography in various ways [66]. While macroscale (>100 μm) patterning mainly affects cells at the colony level, and microtopography, geometry and micropatterning (0.1–100 μm) influence cells at the single-cell level, and nanotopographical features (1.0–100 nm) are able to interact with individual cell receptors [67]. In one study, MC3T3-E1 osteoblasts were cultured on a surface coated with 1D molybdenum selenide (Mo3Se3−) single chain atomic crystals. Compared to control, osteoblast cultures with Mo3Se3− coating demonstrated a significant increase in proliferation (396.2 ± 8.1%). Gold nanoparticles (AuNPs) have also been demonstrated to induce osteogenic differentiation in various studies. One study investigated MSCs cultured on substrates including AuNPs with different sizes, shapes (e.g. sphere, star, nanorod) and diameters with respect to their effect on osteogenic differentiation [68]. It was found that certain AuNP shapes and sizes altered intracellular signaling pathways and increased YAP-mediated osteogenic differentiation. Other substrate materials (e.g. PMMA, polycarbonate, polycaprolactone, Ti, silicon) with engineered topographies created through nanopatterning techniques have been used to evaluate osteogenic differentiation in MSCs and osteoprogenitor cells in vitro in the absence of other osteogenic factors [69]. Pattern types included nanopits, nanopillars, and nanocolumns of varying sizes and dimensions [70]. Overall, the results of these studies suggest that smaller nanopillar features (<20nm in height) and controlled disordered arrangements showed higher osteogenic potential than large nanopillar features (>50nm in height) and ordered arrangements. Alterations in osteogenic differentiation were observed to be generated through RhoA/ROCK and YAP/TAZ-mediated mechanotransduction pathways, as well as direct mechanosensing of the nucleus via the cytoskeleton and the LINC complex.
Engineered materials have also been created that have more complex structures, including hierarchical structures and microarchitectures, in order to more closely mimic the heterogenous physical properties of the extracellular microenvironment in vivo. Material substrates with hierarchical structures have recently been designed to study and regulate cell functions, particularly bone marrow-derived MSCs. Materials with a hierarchical structure have both macro-scale patterns and nano-scale topography, and are thereby considered to more closely resemble the complex extracellular microenvironment of tissues like bone. In a study examining the effects of a hierarchical macropore/nanopore Ti substrate, cultured bone marrow-derived MSCs demonstrated increased development of FAs, increased cytoskeletal tension, and enhanced activity and nuclear translocation of YAP compared to a control substrate. Cells cultured on the material demonstrated increased expression of osteogenic markers ALP, COL I, OPN, OCN, osterix, and decreased expression of the adipogenic markers. In addition, knockdown of YAP inhibited osteogenic differentiation. These findings are consistent with others indicating that mechanotransduction, mediated by YAP, is an important means of hierarchical-structure-induced osteogenic differentiation of bone marrow-derived MSCs [6]. Xie et al. found that MSCs on hierarchical structured Ti coating (HSTC) showed increased cell adhesion and expression of OPN and OCN compared to MSCs on a Ti surface used as control. The authors propose that the increased surface area created by the nanotube layer of HSTC promotes protein adsorption and increased cytoskeletal tension that facilitates osteogenic differentiation [71]. Also, aspects of the hierarchical structure of substrate materials can be tuned to optimize for specific tissue engineering goals, including osteogenic differentiation. Zhou et al. investigated MSC behavior on hierarchical micropore/nanorod-patterned strontium doped hydroxyapatite (Ca9Sr1(PO4)6(OH)2, Sr1-HAP) coatings (MNRs) with interrod spacing size higher than 137 nm. This interrod spacing showed decreased cellular adhesion and proliferation, inhibiting osteogenic differentiation in vitro and in vivo (peri-implant) [72]. However, when the same group examined the impact of nanorod diameter on osteogenic differentiation by utilizing hierarchical MNRs with varying nanorod spacings, they found that the substrate with a nanorod diameter of approximately 70 nm showed the highest increase in osteogenic differentiation via the Wnt/β-catenin pathway. The authors propose that the nanorod diameter of 70 nm was optimal because it is close to the molecular lengths of talin and α-actinin, thereby facilitating the formation of FAs [73].
The supremacy of microscale over nanoscale architecture or vice versa is unclear as both have been demonstrated to effect cell behavior. However, combination of both microscale and nanoscale design achieves multiple levels of organization, allowing for recapitulation of complex hierarchical tissue architecture. An important challenge ahead will be thorough understanding of each individual change on cell biology in a systematic fashion, such that predictions of intracellular signaling may be achieved at the outset of material design. Nevertheless, in skeletal tissues, the necessity for development of hierarchical materials for regeneration continues to lack clarity, as in vivo remodeling and reorganization is expected.
Other materials have been investigated that have bulk properties that are distinct from elements of their microarchitecture. In one study, a polymerized collagen gel was created that had a protein fibrillar microarchitecture (similar to natural ECM) to evaluate its effects on MSC behavior [74]. On gels that had shorter fiber lengths and increased fiber stiffness, MSCs demonstrated a decrease in the transmission of traction forces, leading to the inhibition of cell migration, spreading, and proliferation. This also led MSCs to have decreased formation of FAs, and cells tended to undergo adipogenic differentiation. When collagen density was increased, fiber recruitment and deformed collagen networks contributed to increased cell migration, spreading, and proliferation. These cells had increased FA formation and more often underwent osteogenic differentiation. One group sought to investigate the effects of a 3D substrate material with a macro-porous structure comprised of elastin-like protein (ELP) on MSCs. This macro-porous substrate had some similarities to 2D substrates in that remodeling was not required for cell motility. However, the presentation of matrix stimuli was different because the topography of the pores presented a non-uniform mechanical environment, suggesting differing effects of mechanotransduction [75]. To the authors’ surprise, it was found that higher stiffness of the macro-porous material promoted both osteogenesis and adipogenesis. This result was attributed to differences in cellular orientation and interactions with its substrate material. These data indicate that within porous substrates (like scaffolds), MSC behavior in response to substrate stiffness differs from standard 2D or 3D models and is at least partially determined by the material’s macro-topography.
Ligand Functionalization
Biomolecules, including protein ECM components and other chemical functional groups, can be used to provide specific functionalization of cellular substrate materials. These molecules typically act as anchoring sites that interact with receptors on the cell surface and can be used to induce desired cell-material interactions [50, 76]. These can be used to evaluate the interacting effects of the ligand itself, as well as other mechanical characteristics of the substrate material. In one study, MSCs were grown on PAAm substrates containing collagen I, collagen IV, fibronectin, or laminin to understand the effect of the respective proteins on lineage commitment [77]. While stiffness was still important to osteogenesis, the authors demonstrated that, holding properties constant, the identity of the ECM protein had a significant effect on Runx2 expression. Another study evaluated ligand density as a cue that directly influences mechanotransduction and cell differentiation [78]. In MSCs plated on fibronectin-coated PAAm hydrogels, the translocation of YAP was dependent on substrate stiffness only when ligands were patterned at intermediate densities. At high or low densities, substrate stiffness did not alter YAP localization. It was also found that higher ligand-density led to increased cell spreading, F-actin formation, and osteogenic differentiation (as measured by increased expression of ALP and Runx2) independent of the stiffness of the material. Han et al. examined the impact of lateral spacings of azide terminal ligand nanodomains ranging from 30–60 nm on MSCs cultured on a polystyrene-b-poly(ethylene oxide) (PS–PEO) block copolymers [79]. It was found that on smaller lateral spacings of nanodomains, MSCs showed increased activation of FAK and Src, which led to higher quantities of FAs. FAs on smaller spaced nanodomains showed higher recruitment of myosin IIA and vinculin, thereby allowing those cells to be subjected to higher tension forces than those on larger spacings. This led to increased expression of osteogenic markers including Rac1, cytoplasmic levels of β-catenin, and nuclear translocation of Runx2 and YAP/TAZ. Similar findings were reported by Comisar et al., who instead evaluated MC3T3 pre-osteoblasts plated on RGD (arginine-glycine-aspartic acid) covalently coupled to alginate gels [80].
Metal substrates have also been functionalized in a variety of ways to affect cellular behavior. The physiology of osteoblasts on functionalized metal substrates has been of particular interest given the potential applications for orthopedic implants [81]. For example, in a study evaluating MG-63 osteoblastic cells, Ti was coated with either allylamine plasma polymer (PPAAm) or type-I collagen and compared to an uncoated Ti control [82]. Both functionalized substrates were comparably effective in enhancing FA formation and actin cytoskeleton development. Further electrostatic interactions between PPAAm or type-I collagen and pericellular hyaluronan aided initial steps of osteoblast adhesion to the material, mimicking the function of collagen in the natural ECM [50, 82]. Gold (Au) surfaces can also be functionalized, as they were in a study by Keselowsky and colleagues [83]. Monolayers of alkanethiols were used to modify the Au surface with functional groups (e.g. CH3, OH, COOH, NH2) that had different chemical properties. MC3T3-E1 cells cultured on OH and NH2-terminated surfaces resulted in increases in osteogenesis, which was mediated by binding of β1 integrin. In another study, titanium (Ti) and hydroxyapatite (HAP) were compared as MSC substrate materials in the presence and absence of a collagen overlayer [84]. Ti alone, which had the greatest stiffness, was able to induce the highest levels of myosin II expression in MSCs. However, compared to Ti, hydroxyapatite (HAP) more effectively promoted collagen self-assembly and the formation of the collagen fibrous network, which is critical for MSC motility and osteogenic differentiation. The HAP-collagen overlayer matrix type induced the most favorable stress fiber formation, the longest migration distance (2.8-fold higher than that of pure collagen, and 1.9-fold higher than that of Ti-collagen), and most effectively promoted Wnt/β-catenin mediated osteogenic differentiation. Thus, while the stiffness of substrate material can have a profound effect on cell physiology, the chemical properties of the overlayer material also determine cell processes, behavior, and fate possibly via ligand functionalization. Overall, these and other investigators’ data indicate that modification of metal and inorganic substrate materials can substantially alter cellular signaling pathways involved in osteogenesis.
Mixed and Dynamic Effects
Certain materials, when used as culture substrates, exert their effects on cell physiology due to a complex interplay of multiple properties. Such is the case with graphene, which has the highest elastic modulus of any known substance (0.5–1TPa) and can be applied as a single atom thick later on other surfaces [85]. Despite its stiffness, graphene monolayers only marginally change the elastic modulus of the substance on which they are coated. However, graphene has many ripples and wrinkles on a micrometer scale, is resistive to lateral forces, but has substantial flexibility for out-of-plane deformation. These properties likely facilitate strong anchor points for cells that allow for cytoskeletal tension, thereby leading to mechanotransduction-mediated osteogenesis. In one study by Nayak et al., it was shown that graphene coated on polyethylene terephthalate promotes osteogenesis to an extent comparable to BMP-2[85]. MSCs cultured on graphene displayed significantly increased cell proliferation and osteogenic differentiation compared to those on SiO2 substrate. Similarly, Xie et al. showed that MSCs cultured on PDMS substrates that were coated with a graphene monolayer promoted osteogenic differentiation, without being affected by the intrinsic stiffness of the PDMS material [86]. These cells displayed increased expression of osteogenic proteins including FAK, integrin, Smad1/5, Runx2, and OPN, suggesting that osteogenic differentiation was induced via the integrin/FAK pathway. Graphene has also been studied in the form of a highly porous 3D foam substrate material in order to determine its effects on MSC differentiation and behavior. Crowder et al. found that MSCs cultured on 3D graphene foams strongly promote osteogenic differentiation with increased expression of OPN and OCN, both of which are markers of osteogenesis [87]. Though traditional cell spreading and a total increase in area were not observed, the authors postulate that forced MSC elongation along the bulk of the graphene foam structure (i.e. around/across pores) could be responsible for activation of mechanotransduction pathways, thereby resulting in osteogenesis. Other substrate materials have been created with mixed effects, in addition to other properties like stiffness, depending on their molecular orientation.
Some materials have properties that are dynamic and reversible depending on external stimuli, which ultimately has mixed effects that are distinct from their bulk stiffness. In one study, Wei et al. investigated MSC differentiation on a 3D hydrogel matrix substrate generated with tunable left- or right-handed chirality [88]. Left-handed chirality of the ECM gel was found to increase the clustering of the α5 integrin subunits relative to ECM comprised of the right-handed enantiomer. This was found to lead to increased activation of mechanotransduction, including contractility, FAK and ERK-1/2 cascades, as well as the nuclear translocation of YAP. Another material substrate that has been explored with respect to its dynamic material properties is the conducting polymer Polypyrrole (Ppy). In one study, dynamic switching between nanotips (hydrophilic—poorly adhesive) and nanotubes (hydrophobic—highly adhesive) via a redox process led to modifications in surface adhesion and promoted osteogenic differentiation, regardless of chemical signals or surface stiffness [89]. Cyclic attachment and detachment promoted greater cytoskeleton organization in MSCs cultured on Ppy array. Osteogenic gene expression and nuclear translocation of YAP and Runx2 increased the most with three attachment/detachment cycles. With more cycles, YAP and Runx2 were partly deactivated, likely due to net decreased cell adhesion caused by overstimulation. The aforementioned substrate materials demonstrate how multifactorial the net effects of MSC differentiation are, as they occur in concert with the material’s physical or mechanical properties.
Recently, nanocomposite materials have emerged as a new possibility for tissue engineering and bone regeneration. There has been interest in this relatively new class of biomaterials because natural bone contains a nanocomposite, hierarchical structure with discrete physical and biologic properties. Nanocomposite materials often contain a biodegradable matrix structure and are broadly classifiable as either natural or synthetic polymer based[90]. Some natural polymers include collagen, chitosan, alginate, silk, fucoidan, elastin, gelatin, and HA [91]. Synthetic polymers used include polyethylene glycol, Poly (lactic-co-glycolic) acid, and PLA[92]. These materials also contain nano-scale, bioactive, and easily resorbable fillers designed to impact cell adhesion, proliferation and differentiation [93]. Fabrication can occur via the foam replica method, electrospinning, freeze drying, gas forming, solvent casting/particulate leaching, phase separation, and molecular self-assembly [90, 93, 94]. The nanocomposite matrix structure and its nano-scale filler components act synergistically to promote bone regeneration.
External Mechanical Stimulation
While intrinsic properties of substrates have been shown to modify cell proliferation and differentiation, external stimulation can have a substantial effect on cell behavior and fate of MSCs. Application of external mechanical signals like fluid flow, hydrostatic pressure, compression, and tensile loading, as well as magnetic force, can all modify matrices and/or have a substantial impact on the cells themselves. In addition to the type of stimulation, its frequency and magnitude also have profound effects on cell behavior. Though a full review of the various loading conditions is outside the scope of this review on material substrates, some examples will be covered here. Seo et al. showed that, in 3D gelatin methacryloyl (GelMA) hydrogels, increasing the concentration of hydrogels decreased hMSC spreading while dynamic compression promoted hMSC spreading. It was found that 5% GelMA hydrogels enhanced the sensitivity to compressive strain in hMSCs, and the cells subjected to 42% strain showed the highest increase in osteogenesis, which promoted the expression of osteogenic proteins including Runx2 and OPN. The authors state that this may indicate that 3D hydrogels can lead to increased osteogenic differentiation of cells when stimulated by compression [95]. Zhuang et al. found that application of static magnetic field MC3T3-E1 cells on mineralized collagen coatings (MC) incorporated into iron oxide nanoparticles (IOPs) with outer distributed IOPs (O-IOPs-MC) can lead to increased ALP activity, particularly in cells that were on O-IOP-MC coating with 0.67 IOP-to-collagen mass ratio. These cells also showed increased cell spreading and enhanced expression of genes linked to osteogenesis, including COL-I, ALP, OPN, genes for integrinα1, RhoA, and Runx2. The authors suggest that magnetically deforming collagen coating allows for mechanotransduction and stimulates RhoA-ROCK pathways mediated by integrins, leading MC3T3-E1 cells to undergo osteogenesis [96]. McCoy et al. showed that mechanical stimulation of MSC-seeded 3D collagen glycosaminoglycan scaffolds via a flow-perfusion bioreactor can lead to dose-dependent increases in the expression of placental growth factor (PGF), which was mediated by actin polymerization. Osteogenic differentiation of MSC was dependent on the concentration of PGF, with lower concentrations promoting osteogenesis of MSCs and higher concentrations inducing osteoclastogenesis [8].
Conclusions
The induction of specific cellular responses to materials via mechanotransduction pathways has recently become a robust and growing area of research. Many studies have examined the role of mechanotransduction in determining characteristics and fate of MSCs, including as it relates to osteogenic differentiation and osteogenesis. Numerous materials with different forms and chemical compositions have been engineered as cell culture substrates for this purpose. More recently, there has been growing focus on multifunctional materials that are able influence the development of cultured cells via a number of distinct pathways simultaneously, thereby allowing even more precise control over their fate. This promising new area of research warrants further investigation, potentially leading to substantial improvements in technologies promoting osteogenic regeneration for both scientific and clinical use.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health/National Institute of Dental and Craniofacial Research R01 DE028098 (JCL), the US Department of Veterans Affairs under award number IK2 BX002442, (JCL), the Jean Perkins Foundation (JCL), and the Bernard G. Sarnat Endowment for Craniofacial Biology (JCL).
Footnotes
Disclosures
All authors have no financial interests including products, devices, or drugs associated with this manuscript. There are no commercial associations that might pose or create a conflict of interest with information presented in this submitted manuscript such as consultancies, stock ownership, or patent licensing arrangements. All sources of funds supporting the completion of this manuscript are under the auspices of the University of California Los Angeles.
References
- [1].Cahill KS, Chi JH, Day A, Claus EB, JAMA 2009, 302, 58. [DOI] [PubMed] [Google Scholar]
- [2].Zhu T, Cui Y, Zhang M, Zhao D, Liu G, Ding J, Bioact Mater 2020, 5, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S, Nature 2011, 474, 179. [DOI] [PubMed] [Google Scholar]
- [4].Chen JCJCR, Stem Cell Research & Therapy 2013, 4.23295128 [Google Scholar]
- [5].Martino F, Perestrelo AR, Vinarsky V, Pagliari S, Forte G, Front Physiol 2018, 9, 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pan H, Xie Y, Zhang Z, Li K, Hu D, Zheng X, Fan Q, Tang T, Colloids Surf B Biointerfaces 2017, 152, 344. [DOI] [PubMed] [Google Scholar]
- [7].Morgan JT, Murphy CJ, Russell P, Exp Eye Res 2013, 115, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McCoy RJ, Widaa A, Watters KM, Wuerstle M, Stallings RL, Duffy GP, O’Brien FJ, Stem Cells 2013, 31, 2420. [DOI] [PubMed] [Google Scholar]
- [9].Milloud R, Destaing O, de Mets R, Bourrin-Reynard I, Oddou C, Delon A, Wang I, Albiges-Rizo C, Balland M, Biol Cell 2017, 109, 127. [DOI] [PubMed] [Google Scholar]
- [10].Humphries JD, Byron A, Humphries MJ, J Cell Sci 2006, 119, 3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rahikainen R, von Essen M, Schaefer M, Qi L, Azizi L, Kelly C, Ihalainen TO, Wehrle-Haller B, Bastmeyer M, Huang C, Hytonen VP, Sci Rep 2017, 7, 3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dumbauld DW, Lee TT, Singh A, Scrimgeour J, Gersbach CA, Zamir EA, Fu J, Chen CS, Curtis JE, Craig SW, Garcia AJ, Proc Natl Acad Sci U S A 2013, 110, 9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Uemura A, Nguyen TN, Steele AN, Yamada S, Biophys J 2011, 101, 1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Michael KE, Dumbauld DW, Burns KL, Hanks SK, Garcia AJ, Mol Biol Cell 2009, 20, 2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dupont S, Exp Cell Res 2016, 343, 42. [DOI] [PubMed] [Google Scholar]
- [16].Parsons JT, Horwitz AR, Schwartz MA, Nat Rev Mol Cell Biol 2010, 11, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Naumanen P, Lappalainen P, Hotulainen P, J Microsc 2008, 231, 446; [DOI] [PubMed] [Google Scholar]; Pellegrin S, Mellor H, J Cell Sci 2007, 120, 3491. [DOI] [PubMed] [Google Scholar]
- [18].Hayakawa K, Tatsumi H, Sokabe M, J Cell Biol 2011, 195, 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F, J Cell Biol 2000, 150, 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Engler AJ, Sen S, Sweeney HL, Discher DE, Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- [21].Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS, Nat Methods 2010, 7, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Oka T, Sudol M, Genes Cells 2009, 14, 607. [DOI] [PubMed] [Google Scholar]
- [23].Plouffe SW, Hong AW, Guan KL, Trends Mol Med 2015, 21, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML, Genes Dev 2001, 15, 1229; [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhao W, Li LW, Tian RF, Dong QF, Li PQ, Yan ZF, Yang X, Huo JL, Fei Z, Zhen HN, J Cell Biochem 2019, 120, 17337. [DOI] [PubMed] [Google Scholar]
- [25].Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, Shanahan CM, Trepat X, Navajas D, Garcia-Manyes S, Roca-Cusachs P, Cell 2017, 171, 1397. [DOI] [PubMed] [Google Scholar]
- [26].Fischer M, Rikeit P, Knaus P, Coirault C, Front Physiol 2016, 7, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim NG, Gumbiner BM, J Cell Biol 2015, 210, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lachowski D, Cortes E, Robinson B, Rice A, Rombouts K, Del Rio Hernandez AE, FASEB J 2018, 32, 1099. [DOI] [PubMed] [Google Scholar]
- [29].Sabra H, Brunner M, Mandati V, Wehrle-Haller B, Lallemand D, Ribba AS, Chevalier G, Guardiola P, Block MR, Bouvard D, J Biol Chem 2017, 292, 19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dasgupta I, McCollum D, J Biol Chem 2019, 294, 17693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lin GL, Hankenson KD, J Cell Biochem 2011, 112, 3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Duan P, Bonewald LF, Int J Biochem Cell Biol 2016, 77, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sawakami K, Robling AG, Ai M, Pitner ND, Liu D, Warden SJ, Li J, Maye P, Rowe DW, Duncan RL, Warman ML, Turner CH, J Biol Chem 2006, 281, 23698. [DOI] [PubMed] [Google Scholar]
- [34].Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML, Osteoporosis-Pseudoglioma Syndrome Collaborative G, Cell 2001, 107, 513. [DOI] [PubMed] [Google Scholar]
- [35].Glass DA 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, Karsenty G, Dev Cell 2005, 8, 751. [DOI] [PubMed] [Google Scholar]
- [36].Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA, J Biol Chem 2007, 282, 14515. [DOI] [PubMed] [Google Scholar]
- [37].Kohn AD, Moon RT, Cell Calcium 2005, 38, 439. [DOI] [PubMed] [Google Scholar]
- [38].Uehara S, Udagawa N, Kobayashi Y, Cell Mol Life Sci 2018, 75, 3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S, Cell 2012, 151, 1443. [DOI] [PubMed] [Google Scholar]
- [40].Qiu W, Andersen TE, Bollerslev J, Mandrup S, Abdallah BM, Kassem M, J Bone Miner Res 2007, 22, 1720; [DOI] [PubMed] [Google Scholar]; Arvidson K, Abdallah BM, Applegate LA, Baldini N, Cenni E, Gomez-Barrena E, Granchi D, Kassem M, Konttinen YT, Mustafa K, Pioletti DP, Sillat T, Finne-Wistrand A, J Cell Mol Med 2011, 15, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T, Mol Biol Cell 2003, 14, 2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ghosh-Choudhury N, Abboud SL, Nishimura R, Celeste A, Mahimainathan L, Choudhury GG, J Biol Chem 2002, 277, 33361; [DOI] [PubMed] [Google Scholar]; Li C, Yang X, He Y, Ye G, Li X, Zhang X, Zhou L, Deng F, Int J Med Sci 2012, 9, 862; [DOI] [PMC free article] [PubMed] [Google Scholar]; Lou J, Tu Y, Li S, Manske PR, Biochem Biophys Res Commun 2000, 268, 757. [DOI] [PubMed] [Google Scholar]
- [43].Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massague J, Cell 2009, 139, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, Wrana JL, Attisano L, Dev Cell 2010, 18, 579; [DOI] [PubMed] [Google Scholar]; Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL, Dev Cell 2010, 19, 831. [DOI] [PubMed] [Google Scholar]
- [45].Rawadi G, Vayssière B, Dunn F, Baron R, Roman-Roman S, J Bone Miner Res 2003, 18, 1842. [DOI] [PubMed] [Google Scholar]
- [46].Yan Y, Tang D, Chen M, Huang J, Xie R, Jonason JH, Tan X, Hou W, Reynolds D, Hsu W, Harris SE, Puzas JE, Awad H, O’Keefe RJ, Boyce BF, Chen D, J Cell Sci 2009, 122, 3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mbalaviele G, Sheikh S, Stains JP, Salazar VS, Cheng SL, Chen D, Civitelli R, J Cell Biochem 2005, 94, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen Y, Whetstone HC, Youn A, Nadesan P, Chow EC, Lin AC, Alman BA, J Biol Chem 2007, 282, 526. [DOI] [PubMed] [Google Scholar]
- [49].Kokabu S, Rosen V, FEBS Open Bio 2018, 8, 168; [DOI] [PMC free article] [PubMed] [Google Scholar]; Im J, Kim H, Kim S, Jho EH, Biochem Biophys Res Commun 2007, 354, 296. [DOI] [PubMed] [Google Scholar]
- [50].Jang YH, Jin X, Shankar P, Lee JH, Jo K, Lim KI, Int J Mol Sci 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Xing Q, Yates K, Vogt C, Qian Z, Frost MC, Zhao F, Sci Rep 2014, 4, 4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Stutchbury B, Atherton P, Tsang R, Wang DY, Ballestrem C, J Cell Sci 2017, 130, 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Witkowska-Zimny M, Wrobel E, Mrowka P, Folia Histochem Cytobiol 2014, 52, 297. [DOI] [PubMed] [Google Scholar]
- [54].Zhou C, Zhang D, Zou J, Li X, Zou S, Xie J, ACS Appl Mater Interfaces 2019, 11, 26448. [DOI] [PubMed] [Google Scholar]
- [55].Liu N, Zhou M, Zhang Q, Zhang T, Tian T, Ma Q, Xue C, Lin S, Cai X, Cell Prolif 2018, 51, e12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Khatiwala CB, Peyton SR, Metzke M, Putnam AJ, J Cell Physiol 2007, 211, 661. [DOI] [PubMed] [Google Scholar]
- [57].Khatiwala CB, Kim PD, Peyton SR, Putnam AJ, J Bone Miner Res 2009, 24, 886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS, Dev Cell 2004, 6, 483. [DOI] [PubMed] [Google Scholar]
- [59].Parekh SH, Chatterjee K, Lin-Gibson S, Moore NM, Cicerone MT, Young MF, Simon CG Jr., Biomaterials 2011, 32, 2256; [DOI] [PMC free article] [PubMed] [Google Scholar]; Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ, Nat Mater 2010, 9, 518; [DOI] [PMC free article] [PubMed] [Google Scholar]; Pek YS, Wan AC, Ying JY, Biomaterials 2010, 31, 385; [DOI] [PubMed] [Google Scholar]; Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A, Li S, Biomaterials 2011, 32, 3921; [DOI] [PMC free article] [PubMed] [Google Scholar]; Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA, Nat Mater 2013, 12, 458; [DOI] [PMC free article] [PubMed] [Google Scholar]; Evans ND, Minelli C, Gentleman E, LaPointe V, Patankar SN, Kallivretaki M, Chen X, Roberts CJ, Stevens MM, Eur Cell Mater 2009, 18, 1; [DOI] [PubMed] [Google Scholar]; Zhang T, Lin S, Shao X, Zhang Q, Xue C, Zhang S, Lin Y, Zhu B, Cai X, Cell Prolif 2017, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cross LM, Shah K, Palani S, Peak CW, Gaharwar AK, Nanomedicine 2018, 14, 2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Razafiarison T, Holenstein CN, Stauber T, Jovic M, Vertudes E, Loparic M, Kawecki M, Bernard L, Silvan U, Snedeker JG, Proc Natl Acad Sci U S A 2018, 115, 4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Werner M, Blanquer SB, Haimi SP, Korus G, Dunlop JW, Duda GN, Grijpma DW, Petersen A, Adv Sci (Weinh) 2017, 4, 1600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kilian KA, Bugarija B, Lahn BT, Mrksich M, Proc Natl Acad Sci U S A 2010, 107, 4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee MS, Lee DH, Jeon J, Oh SH, Yang HS, ACS Appl Mater Interfaces 2018, 10, 38780. [DOI] [PubMed] [Google Scholar]
- [65].Lee JH, Lee YJ, Cho HJ, Shin H, Tissue Eng Part A 2014, 20, 2031. [DOI] [PubMed] [Google Scholar]
- [66].Lee JW, Chae S, Oh S, Kim SH, Choi KH, Meeseepong M, Chang J, Kim N, Yong Ho K, Lee NE, Lee JH, Choi JY, Nano Lett 2018, 18, 7619; [DOI] [PubMed] [Google Scholar]; Gentile F, Tirinato L, Battista E, Causa F, Liberale C, di Fabrizio EM, Decuzzi P, Biomaterials 2010, 31, 7205; [DOI] [PubMed] [Google Scholar]; Lacerda L, Bianco A, Prato M, Kostarelos K, Adv Drug Deliv Rev 2006, 58, 1460; [DOI] [PubMed] [Google Scholar]; Garcia-Gradilla V, Sattayasamitsathit S, Soto F, Kuralay F, Yardimci C, Wiitala D, Galarnyk M, Wang J, Small 2014, 10, 4154. [DOI] [PubMed] [Google Scholar]
- [67].Dalby MJ, Gadegaard N, Oreffo RO, Nat Mater 2014, 13, 558. [DOI] [PubMed] [Google Scholar]
- [68].Li J, Li JJ, Zhang J, Wang X, Kawazoe N, Chen G, Nanoscale 2016, 8, 7992. [DOI] [PubMed] [Google Scholar]
- [69].Dobbenga S, Fratila-Apachitei LE, Zadpoor AA, Acta Biomater 2016, 46, 3. [DOI] [PubMed] [Google Scholar]
- [70].Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CD, Oreffo RO, Nat Mater 2007, 6, 997; [DOI] [PubMed] [Google Scholar]; Lavenus S, Berreur M, Trichet V, Pilet P, Louarn G, Layrolle P, Eur Cell Mater 2011, 22, 84; [DOI] [PubMed] [Google Scholar]; McNamara LE, Sjostrom T, Burgess KE, Kim JJ, Liu E, Gordonov S, Moghe PV, Meek RM, Oreffo RO, Su B, Dalby MJ, Biomaterials 2011, 32, 7403; [DOI] [PubMed] [Google Scholar]; Guvendik S, Trabzon L, Ramazanoglu M, J Nanosci Nanotechnol 2011, 11, 8896. [DOI] [PubMed] [Google Scholar]
- [71].Xie Y, Ao H, Xin S, Zheng X, Ding C, Mater Sci Eng C Mater Biol Appl 2014, 38, 272. [DOI] [PubMed] [Google Scholar]
- [72].Zhou J, Li B, Han Y, Zhao L, Nanomedicine 2016, 12, 1161. [DOI] [PubMed] [Google Scholar]
- [73].Zhou J, Zhao L, Li B, Han Y, Nanomedicine 2018, 14, 1719. [DOI] [PubMed] [Google Scholar]
- [74].Xie J, Bao M, Bruekers SMC, Huck WTS, ACS Appl Mater Interfaces 2017, 9, 19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Haugh MG, Vaughan TJ, Madl CM, Raftery RM, McNamara LM, O’Brien FJ, Heilshorn SC, Biomaterials 2018, 171, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hackett AJ, Malmström J, Travas-Sejdic J, Progress in Polymer Science 2017, 70, 18; [Google Scholar]; Hoshiba T, Tanaka M, Anal Sci 2016, 32, 1151. [DOI] [PubMed] [Google Scholar]
- [77].Rowlands AS, George PA, Cooper-White JJ, Am J Physiol Cell Physiol 2008, 295, C1037. [DOI] [PubMed] [Google Scholar]
- [78].Stanton AE, Tong X, Lee S, Yang F, ACS Appl Mater Interfaces 2019, 11, 8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Han P, Frith JE, Gomez GA, Yap AS, O’Neill GM, Cooper-White JJ, ACS Nano 2019, 13, 11129. [DOI] [PubMed] [Google Scholar]
- [80].Comisar WA, Kazmers NH, Mooney DJ, Linderman JJ, Biomaterials 2007, 28, 4409. [DOI] [PubMed] [Google Scholar]
- [81].Rammelt S, Illert T, Bierbaum S, Scharnweber D, Zwipp H, Schneiders W, Biomaterials 2006, 27, 5561; [DOI] [PubMed] [Google Scholar]; Petrie TA, Capadona JR, Reyes CD, Garcia AJ, Biomaterials 2006, 27, 5459; [DOI] [PubMed] [Google Scholar]; Elmengaard B, Bechtold JE, Soballe K, J Biomed Mater Res A 2005, 75, 249. [DOI] [PubMed] [Google Scholar]
- [82].Finke B, Luethen F, Schroeder K, Mueller PD, Bergemann C, Frant M, Ohl A, Nebe BJ, Biomaterials 2007, 28, 4521. [DOI] [PubMed] [Google Scholar]
- [83].Keselowsky BG, Collard DM, Garcia AJ, Proc Natl Acad Sci U S A 2005, 102, 5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].He J, Meng G, Yao R, Jiang B, Wu Y, Wu F, J Mech Behav Biomed Mater 2016, 59, 353. [DOI] [PubMed] [Google Scholar]
- [85].Nayak TR, Andersen H, Makam VS, Khaw C, Bae S, Xu X, Ee PL, Ahn JH, Hong BH, Pastorin G, Ozyilmaz B, ACS Nano 2011, 5, 4670. [DOI] [PubMed] [Google Scholar]
- [86].Xie H, Cao T, Franco-Obregon A, Rosa V, Int J Mol Sci 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Crowder SW, Prasai D, Rath R, Balikov DA, Bae H, Bolotin KI, Sung HJ, Nanoscale 2013, 5, 4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wei Y, Jiang S, Si M, Zhang X, Liu J, Wang Z, Cao C, Huang J, Huang H, Chen L, Wang S, Feng C, Deng X, Jiang L, Adv Mater 2019, 31, e1900582. [DOI] [PubMed] [Google Scholar]
- [89].Wei Y, Mo X, Zhang P, Li Y, Liao J, Li Y, Zhang J, Ning C, Wang S, Deng X, Jiang L, ACS Nano 2017, 11, 5915. [DOI] [PubMed] [Google Scholar]
- [90].Bharadwaz A, Jayasuriya AC, Mater Sci Eng C Mater Biol Appl 2020, 110, 110698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Saravanan S, Leena RS, Selvamurugan N, Int J Biol Macromol 2016, 93, 1354. [DOI] [PubMed] [Google Scholar]
- [92].Rao SH, Harini B, Shadamarshan RPK, Balagangadharan K, Selvamurugan N, Int J Biol Macromol 2018, 110, 88. [DOI] [PubMed] [Google Scholar]
- [93].Pina S, Oliveira JM, Reis RL, Adv Mater 2015, 27, 1143. [DOI] [PubMed] [Google Scholar]
- [94].Oliveira JM, Silva SS, Malafaya PB, Rodrigues MT, Kotobuki N, Hirose M, Gomes ME, Mano JF, Ohgushi H, Reis RL, J Biomed Mater Res A 2009, 91, 175; [DOI] [PubMed] [Google Scholar]; Chae T, Yang H, Leung V, Ko F, Troczynski T, J Mater Sci Mater Med 2013, 24, 1885; [DOI] [PubMed] [Google Scholar]; Dehghani F, Annabi N, Curr Opin Biotechnol 2011, 22, 661; [DOI] [PubMed] [Google Scholar]; Hou Q, Grijpma DW, Feijen J, Biomaterials 2003, 24, 1937; [DOI] [PubMed] [Google Scholar]; Matson JB, Zha RH, Stupp SI, Curr Opin Solid State Mater Sci 2011, 15, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Seo J, Shin JY, Leijten J, Jeon O, Bal Ozturk A, Rouwkema J, Li Y, Shin SR, Hajiali H, Alsberg E, Khademhosseini A, ACS Appl Mater Interfaces 2018, 10, 13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Zhuang J, Lin S, Dong L, Cheng K, Weng W, Acta Biomater 2018, 71, 49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.