Abstract
Immune reconstitution following allogeneic hematopoietic stem cell transplantation (allo-HSCT) sets the stage for the goal of a successful transplant—the prevention of disease relapse without graft versus host disease (GVHD) and opportunistic infection. In both epidemiologic studies and in controlled animal studies, it is known that the gut microbiome (GM) can profoundly influence normal innate and adaptive immune development and can be altered by microbial transfer and antibiotics. Following allo-HSCT the GM has been shown to influence clinical outcomes but published associations between the GM and immune reconstitution post-allo-HSCT are lacking. In this viewpoint we propose that the extensive knowledge garnered from studying normal immune development can serve as a framework for studying immune development post-allo-HSCT. We summarize existing studies addressing the effect of the GM on immune ontogeny and draw associations with immune reconstitution and the GM post-allo-HSCT.
Keywords: microbiome, allogeneic, transplant, metagenomics, immune reconstitution
INTRODUCTION
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a curable modality for many hematologic malignancies and bone marrow disorders [1]. Following conditioning chemotherapy, donor HSCs and co-infused donor immune cells establish new innate and adaptive immune systems that are anticipated to protect the host from infection and relapse—the latter referred to as a graft versus malignancy (GvM) effect [2,3]. Immune reconstitution, however, carries the risk of graft versus host disease (GvHD)—a multisystem acute and/or chronic immune-mediated disease, characterized by donor cell reactivity to host tissues [4]. Immunosuppressive therapies (IST) have improved GvHD-related mortality [1]; however, the ability to consistently eliminate GvHD without impacting GvM and infection risk is lacking. The gut microbiome (GM) has been shown to fine-tune elements of the developing immune system in infancy, resulting in lasting impacts on immunity throughout life [5]. Emerging evidence is now revealing that the GM can also impact immune development post-allo-HSCT in murine models and in patients [6–8]. We review the role the GM plays in shaping neonatal immune development, and how this can serve as a framework that will guide further investigations into how GM signals alter immunity post-allo-HSCT.
THE IMPORTANCE OF POST-ALLO-HSCT IMMUNE RECONSTITUTION KINETICS
Immune reconstitution post-allo-HSCT occurs over approximately two years, encompassing the recovery of both effector and regulatory immune cell subsets (Figure 1). For an in-depth discussion of the impacts of donor and host variables on the kinetics of immune recovery the reader is referred to excellent recent reviews [3,9]. Studies assessing the impact of the magnitude and speed of immune recovery on allo-HSCT outcomes are mostly retrospective cohort analyses from single centers and are thus subject to variations in clinical practice, such as conditioning regimens, HSC dose, GVHD prophylaxis, ex vivo HSC manipulation [3,10,11]. Nevertheless, interesting insights into recovery of adaptive and innate immunity have been obtained and are summarized in Table 1.
Figure 1. Time to recovery of immune cell subsets following allogeneic hematopoietic stem cell transplantation (allo-HSCT) and representative associations between components of the gut microbiome and selected immune subsets in normal development.
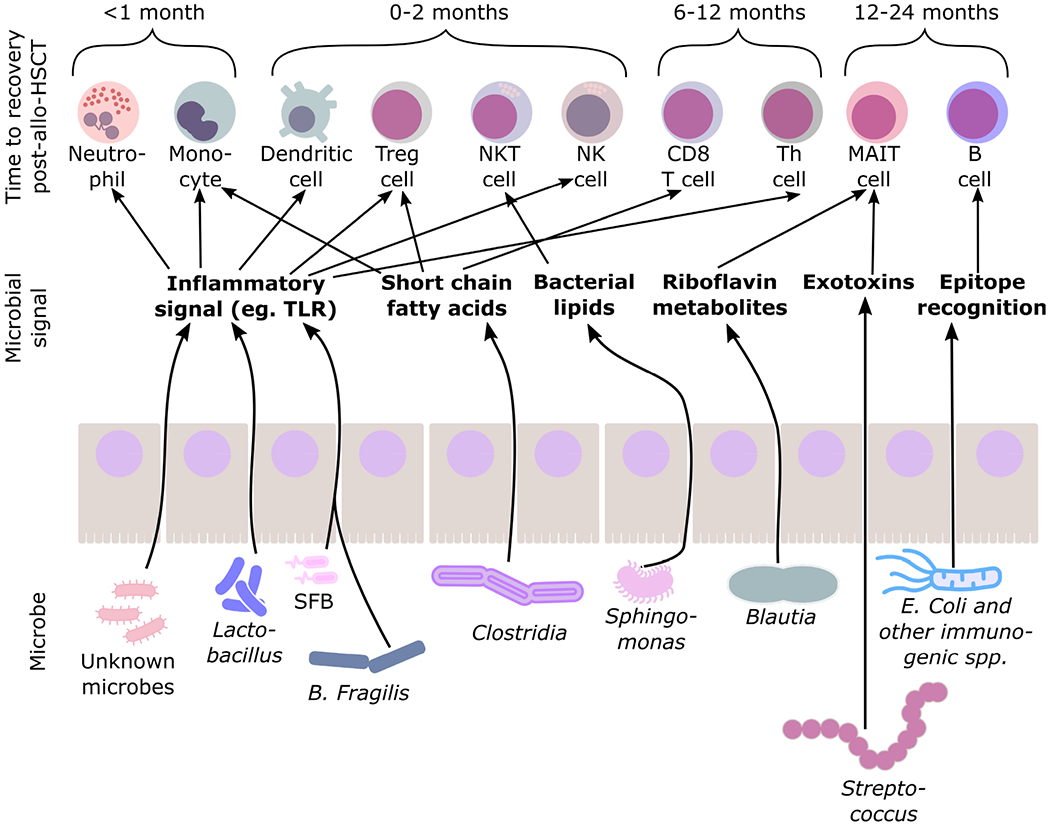
Abbreviations: NK, natural killer; Th, T helper cells; Treg, Regulatory T cell; MAIT, mucosal-associated invariant T cell; SFB, segmented filamentous bacteria. References as per Table 2.
Table 1.
Association of immune cell subsets with relapse, infection and acute graft versus host disease from patient data.
| Immune cell subset | Effect on GVM and infection [refs] | Effect on acute GVHD [refs] |
|---|---|---|
| Neutrophil | Early recovery has no clear effect on GVM. Protects against opportunistic infection [30] | Early recovery associated with increased risk of GVHD [30] |
| Monocyte | Early recovery associated with less relapse in certain subsets [31] | Timing of recovery not clearly associated with aGVHD [15] |
| Dendritic cell | Low plasmacytoid dendritic cell recovery associated with relapse and infection [32] | High plasmacytoid and myeloid dendritic cell recovery associated with less incidence of severe aGVHD [32,33] |
| NK cell | Early recovery associated with less relapse [13,14] | Early recovery associated with increased risk of aGVHD [13,14] |
| NKT Cell | Early recovery associated with less relapse [3,24,29] | Early recovery associated with increased risk of aGVHD [3,24,29] |
| γδ T cell | Early recovery associated with less relapse and infection [26–28] | No Effect on aGVHD [28] |
| MAIT cell | No clear association with infection or relapse [3] | Early recovery associated with less GVHD [25,34] |
| CD8+ T cell | Early recovery associated with less relapse and infection [19–22] | Early recovery associated with increased risk of aGVHD [19–22] |
| CD4+ Th cell | Subset dependent. Mostly associated with decreased risk of infection and relapse. [3,21] | Subset dependent. Mostly associated with increased risk of aGVHD [3] |
| CD4+ Treg cell | Early recovery may be associated with increased risk of relapse [33] | Higher cell numbers associated with less aGVHD [3] |
| B cell | Early recovery associated with less infection [14,29] | Early recovery associated with decreased risk of aGVHD [29] |
Abbreviations: aGVHD, acute graft versus host disease; NK, natural killer; Th, T helper cell; Treg, T regulatory cell.
Enhanced neutrophil recovery provides innate immune defense against bacterial and fungal infections and can shorten hospital stay, yet has been associated with a risk of potentiating GvHD [12]. Likewise, increased natural killer (NK) cell recovery at days 28 and 56 decreases the risk of relapse, but may come at the expense of increased risk of GVHD [13,14]. Although early monocyte recovery is associated with decreased risk of relapse and improved overall survival, [15–17] early recovery of immune regulatory myeloid derived-suppressor cells at day 14 increases the risk of both relapse and paradoxically severe acute GVHD [18].
Within the adaptive immune system, the extent and timing of recovery of distinct T cell subsets has important clinical implications. Early CD8+ T cell recovery is associated with decreased relapse risk and viral infection, but increased risk of acute GVHD [19–22]. Early reconstitution of CD45RA− CD62L− regulatory T cells (Tregs) at 3 months and long-lived, CD45RA+ CD62L+ Tregs at 6 months is associated with decreased incidence of chronic GVHD [23,24]. Rapid reconstitution of non-conventional T cells may influence the risk of post-transplant opportunistic infections (in the case of γδ T cells), GVHD (NKT and mucosal-associated invariant T, MAIT cells), and relapse (γδ and NKT cells) [25–28]. Early B cell recovery is associated with superior anti-viral, anti-bacterial immunity and response to vaccination and decreased non-relapse mortality [14,29]. Thus, the kinetics of reconstitution of numerous distinct immune subsets contribute to the outcomes of allo-HSCT.
THE GM AND IMMUNE DEVELOPMENT
Since the association between GM composition in childhood and autoimmune disease in later life was established in the 1990s [35,36] there have been intense investigations into the mechanisms by which the GM composition impacts development of adaptive and innate immune cell stimulation and development in both mice and human cells in culture (Figure 1 and Table 2). By comparing mice raised in completely germ free (GF) environments to mice exposed to known microbes (gnotobiotic), or against mice harboring an array of normal microbes but lacking in certain pathogens (specific-pathogen-free, SPF) with or without antibiotic exposure, multiple studies have demonstrated the impact of the GM on specific components of immune development and subsequent immunity [37]. For example, absolute T cell and B cell counts are similar between SPF and GF mice, but CD4+ T-helper (Th) cell numbers are lower and Th2-biased cells higher in GF mice—a phenotype that can be rescued in GF mice by oral administration of bacterial polysaccharide (PSA) from the gut commensal Bacteroides fragilis [38]. PSA signals via toll-like receptor 2 (TLR2) to thymic-migrating dendritic cells (DCs), which can regulate PLZF-dependent transcriptional programs in developing thymic T cells, resulting in protection against autoimmune colitis and fatal inflammation from herpes viral encephalitis [38–40]. Segmented filamentous bacteria adhere to epithelial cells of the gut and in doing so create an inflammatory environment that supports the development Th17 cells that in turn cause ulcerative colitis in mouse models, which can be mitigated by the transplantation of Treg activating GM species [41,42]. Other pro-inflammatory bacterial components, such as exotoxins from Group A Streptococcus, have been shown to augment Treg proliferation in human cells in vitro and function through a monocyte dependent-mechanism thereby protecting against autoimmune disease in mouse models; however, it remains to be seen if this phenomenon is related to an initial inflammatory response or represents a distinct developmental program [43].
Table 2.
Examples of the impact of gut microbiota on immune cell development in mice and human in vitro model systems.
| Immune cell | Model system [ref] | Bacteria | Bacterial component & mechanism |
|---|---|---|---|
| Neutrophil | Antibiotic-treated mice [44] | Loss of: Proteobacteria (early); Firmicutes, class: Bacilli (day 5–14); Bacteroidetes, class: Bacteroidia (late) | ↓ PAMP → ↓ stimulation of ILC → ↓ GCSF → neutrophils |
| Antibiotic-treated allo-HSCT mice [6] | ↓ bacterial load and diversity | ↓ bacterial metabolism → ↓ digested sugars → ↓ neutrophils | |
| Monocyte | Antibiotic-treated mice [45] | Loss of Lactobacillus | ↓ SCFA → ↓ monocyte activation |
| Human U937 monoblastic leukemia cell line [46] | SCFA producing bacteria (phyla Firmicutes and Bacteroidetes) | ↓ SCFA → ↓ inflammasome complex → ↓ monocyte activation | |
| Stimulated human healthy donor cells [43] | Streptococcus pyogenes | ↓ exotoxin → ↓ monocyte stimulation and differentiation | |
| Dendritic cell | Gnotobiotic GF and comparison with SPF mice [38] | Bacteroides fragilis | Polysaccharide → ↑ TLR2 activation → plasmacytoid DC development |
| Antibiotic-treated mice [47] | Lactobacillus plantarum | Glycolipid → mincle receptor on DC → DC activation | |
| NK cell | Lactobacillus treated healthy human donor cells [48] | Bifidobacterium bifidum and Lactobacillus reuteri | Stimulation of DC → activation and expansion of NK cells |
| NKT Cell | Gnotobiotic GF mice [49] | Sphingomonas spp. | Bacterial lipids → bind to CD1d → NKT stimulation and expansion |
| Comparison of GF and SPF mice [39] | Bacteroides fragilis | Polysaccharide → ↑ TLR2 activation → DC migration from gut to thymus → thymic iNKT development | |
| MAIT cell | Comparison of GF and SPF mice [50] | Riboflavin metabolizing bacteria (e.g., Blautia spp.) | 5-ARU → 5-OPRU → bound to MR1 → MAIT cell stimulation and expansion |
| Healthy human donor cells stimulated by MR1 bound riboflavin metabolites [51] | (No bacteria used) | MR1 bound riboflavin metabolites → MAIT cell stimulation and expansion | |
| CD8+ T cell | Comparison of GF and SPF mice [52] | Specific bacteria not identified | Microbial-derived butyrate → ↓ glycolysis and ↑ oxidative phosphorylation → promotes memory formation from activated T cells |
| CD4+ Th cell | Gnotobiotic GF and comparison with SPF mice [38] | Bacteroides fragilis | Polysaccharide → ↑ TLR2 activation → plasmacytoid DC development → ↑ Th cell |
| Treatment of SPF mice with bacteria | Bacteroides fragilis | Polysaccharide → binds to B cells → CD4+ Th development | |
| Gnotobiotic GF | Segmented filamentous bacteria | Adherence to mucosa → SAA and ROS → TH17 development | |
| CD4+ Treg cell | Gnotobiotic GF mice [53] | Clostridioides difficile | Butyrate production → epigenomic change at FoxP3 locus → ↑ Treg development |
| B cell | Gnotobiotic GF mice [54,55] | Escherichia coli K12 variant | High bacterial load in gut → ↑ B cell IgA repertoire |
Abbreviations: PAMP, pathogen association molecular pattern; GCSF, granulocyte colony stimulating factor; allo-HSCT, allogeneic hematopoietic stem cell transplantation; SCFA, short chain fatty acid; GF, germ free; SPF, specific-pathogen-free; TLR, toll-like receptor; DC, dendritic cell; NK, natural killer; 5-ARU, 5-amino-6-d-ribitylaminouracil; 5-OPRU, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil; MAIT, mucosal-associated invariant T cell; Th, T helper cell; Treg, T regulatory cell; SFB, segmented filamentous bacteria.
B cell development, and the immunoglobulin A (IgA) repertoire in particular, is shaped by sampling and presentation of GM antigens by subepithelial mesenchymal cells and dendritic cells within mucosal-associated lymphoid tissue [56]. Unlike systemic bacterial immunization which drives IgG-mediated responses with only low bacterial numbers, high bacterial loads are required for the development of antigen-specific IgA-mediated immunity [54,55]. These distinct thresholds and requirements for constant sampling and presentation allow directed polyclonal IgA responses in the face of antigenic diversity within the GM, whilst somatic hypermutation within the B cell compartment allows diversification of the IgA-secreting repertoire [55,57,58].
Metabolic byproducts of the gut microbiome can influence development of distinct T cell subsets. MAIT cells express an invariant T cell receptor that is responsive to riboflavin metabolites unable to be synthesized by animals but produced by diverse pathogenic and commensal GM organisms. These riboflavin metabolites are presented to MAIT cells by the major-histocompatibility related protein 1 (MR1), resulting in MAIT cell activation [51]. Loss of MR1 exacerbates GVHD in the gut in animal models [59]. Similarly, bacterial lipids which bind to CD1d on gut dendritic cells can potentiate NKT cell development and activation [49]. Butyrate-producing bacteria metabolizing host bile acids alter epigenomic acetylation of the FoxP3 locus leading to Treg development that can be co-opted to protect against autoimmune disease in GF mice; however, this also impairs anti-tumor responses [53,60]. Although absolute CD8+ T cell counts are not altered in GF environments, microbial-derived butyrate promotes memory T cell activation and survival by subverting glycolysis and directly promoting fatty acid use in oxidative phosphorylation [52].
Short chain fatty acids can drive innate immunity by activating human macrophage cell lines thereby promoting local gut immunity [46]. Pathogen-associated molecular patterns (PAMPs) for GM bacteria also directly stimulate the differentiation and growth of dendritic cells and potentiate protective immunity in non-GI organs [47]. In antibiotic-treated neonatal mice decolonized of commensal bacteria, stimulatory microbial signals to innate lymphoid cells (ILCs) are lacking, resulting in decreased IL-17 (IL-17) and granulocyte colony stimulating factor (G-CSF) production with subsequent impaired granulopoiesis and susceptibility to Escherichia coli sepsis [44]. Impaired ILC type 3 development in antibiotic exposed neonatal mice also directly impairs mucosal immunity and increases susceptibility to Klebsiella pneumoniae [61].
ALTERATIONS IN THE GM FOLLOWING ALLO-HSCT
From species identification by 16S rRNA sequencing (16Sseq) of stool it is known that GM bacterial diversity in adult and pediatric patients undergoing allo-HSCT is less than that seen in healthy controls and may take up to five months to recover [62–64]. At a species-specific level, in children there is a shift away from microbes commonly associated with healthy GM, towards more pathogenic genera such as Staphylococcus and Enterobacter. Likewise, in adults a dysbiotic shift towards Enteroccocus within phylum Firmicutes and an increase in obligate anaerobes is also observed [62,64,65]. The reasons underlying these marked post-allo-HSCT changes in GM composition are complex and interacting. Exposure to antibiotics, radiation and chemotherapy prior to transplant can be directly toxic to some GM species, and conditioning therapy often disrupts mucosal barriers and impairs mucosal immunity [66,67]. Disruption of the intestinal barrier by radiation, conditioning chemotherapy or gut GVHD may lead to loss of Paneth cells with subsequent decrease secretion of defensins and overgrowth or normally defensin-sensitive species [68]. The loss of the intestinal mucosal barrier may also exacerbate GVHD and lead to a reliance on total parenteral nutrition which in turn is associated with dysbiosis [69].
Technological and statistical limitations may also limit a complete appreciation of the GM post-allo-HSCT and subsequent functional implications of dysbiosis. Clustering of 16Sseq data as operational taxonomic units (OTUs) allows classification and grouping of similar sequences as OTUs, but usually limits bacterial assignment to genera, thus eschewing species-specific effects. This potentially dilutes competing effects of species within the same genus; and often does not address the contribution of the virome and fungome [70,71]. Furthermore, bacteria exist in communities, share genetic elements, and display metabolic cooperativity that cannot be comprehensively captured by taxonomic identification [72]. New approaches, such as shotgun metagenomic sequencing (SMGS) allow identification of individual bacterial, viral and fungal species and allow associations at the gene-level, providing an opportunity for more complete and mechanistic associations of the GM with clinical outcomes yet are contingent on the ever-increasing annotation of microbial genomes [73,74]. Creating associations between GM and transplant outcomes from large datasets may also require the use of novel, iterative statistical approaches that are dependent on large-scale computing power and take into account prior probabilities, such as sequencing depth, as well as draw associations not only between total read count of genes but variations in reads [75,76]. Such Bayesian and frequentist methods are already being implemented as outlined in the subsequent section [7,8].
ASSOCIATION OF THE GM WITH POST-ALLO-HSCT OUTCOMES AND IMMUNE RECONSTITUTION
Empirical Evidence from Murine Model Systems
The influence of the GM on allo-HSCT outcomes is most clearly seen in antibiotic-treated SPF and gnotobiotic mouse models where the GM composition can be controlled [77]. Innate immune cells activated by gut dysbiosis through pathogen sensing TLRs produce reactive oxygen species that can mediate tissue injury in mouse GVHD [78]. Co-housing of transplanted wild-type (WT) mice with IL-17−/− mice susceptible to hyperacute GVHD, results in altered GM in WT mice and subsequent increased aGVHD susceptibility [79]. The post-allo-HSCT GM has also been shown to increase IL-12/23p40 subunit liberation that upregulates MHC-II on intestinal epithelial cells and initiates lethal GVHD [80]. Lactose excess has been shown to drive Enterococcal domination that is associated with aGVHD, whilst microbial liberation of indoles elicits a type 1 interferon signal, paradoxically shown to protect against aGVHD [67,81].
Despite the mechanistic associations of immune development with microbial metabolism in the gut and the marked alterations in gut microbiota following allo-HSCT, few reports have drawn direct links between the gut microbiome and post-allo-HSCT immune reconstitution in mouse models. Staffas at al. demonstrated that antibiotic-treated mice showed decreased granulocyte and lymphocyte recovery following allo-HSCT [6]. In a series of elegant experiments it was further shown that liberation of sucrose from gut microbiota post-allo-HSCT stimulates granulopoiesis and that sucrose supplementation can be used as a post-biotic to improve neutrophil recovery in antibiotic treated mice [6]. Previous studies have also demonstrated that GF and antibiotic-treated mice show reduced HSC and thus impaired normal granulopoiesis and lymphopoiesis [82,83]. However, the study by Staffas et al. showed that HSC numbers were relatively unaffected indicating that the GM may alter more differentiated progenitor cells after allo-HSCT compared to normal immune development [6]. This hypothesis comes with the caveat that these studies differed in the use of antibiotic-treated mice versus GF mice and the regimen of antibiotics used.
Clinical Associations in Patients
In adult patients, associations between GM and allo-HSCT outcomes rely on temporal delineations between gut dysbiosis, immune reconstitution and transplant outcome or therapy-induced alterations in the GM that result in clinical outcomes. In the largest study to date, van den Brink and colleagues showed in a multi-center study comprising 1362 patients at four centers that microbial diversity in the peri-engraftment, but not pre-engraftment, period is associated with overall survival when controlling for antibiotic exposure [71]. Interestingly, the effect of stool microbial diversity on overall survival was only seen in the subset of patients who received T cell replete grafts implying an interaction between T cell immune reconstitution and the GM [71]. In a recent follow-up study from the same center, Bayesian regression was used to show decreased microbial diversity was associated with impaired neutrophil and lymphocyte recovery [8]. Corroborating results seen in mice undergoing allo-HSCT [6], it was further shown that Ruminococcus 2 species were associated improved lymphocyte recovery [78]. Further reports from this large cohort detailing exact immune subsets associated with recovery are awaited.
Other single center studies have shown an association between incidence and severity of aGVHD and subsequent mortality and reduced GM diversity in the peri-engraftment period with only one study documenting an association of GVHD with pre-transplant GM composition [64,84–90]. Confirming these results, antibiotic exposure both and pre- and post-allo-HSCT decreases GM diversity and increases the risk of GVHD [66,77,91]. Dysbiosis and bacterial predominance within the GM can also lead to translocation and subsequent blood stream infection with attendant morbidity and mortality, even with species not commonly associated with gut colonization such as S. epidermidis and P. aeruginosa [92]. The association of species loss and increased risk of GVHD is further corroborated by evidence of the utility of fecal microbiota transplantation (FMT) to increase GM diversity and thus treat even steroid-refractory acute GVHD whilst also enhancing monocyte and lymphocyte recovery [8,93–96].
In pediatric populations there is also evidence of the interaction between the post-allo-HSCT GM and transplant outcomes, yet with only one study to data reporting associations with immune reconstitution. Antibiotic exposure is associated with decreased commensal species of the Clostridial class and subsequent increased risk of gut GVHD in children post-allo-HSCT [97]. Other pediatric studies have confirmed results seen in adult patients showing an association of GVHD with a loss of Blautia spp. [71,98].
In a comprehensive, data-dense study, Ingham et al. undertook longitudinal monitoring of immune recovery, markers of inflammation and GM composition in 37 children prior to and following allo-HSCT and confirm that in children microbial diversity markedly decreases following allo-HSCT and did not return to pre-allo-HSCT levels after 5 weeks. Using multivariate regression analysis with sparse partial least squares regression (sPLS) followed by hierarchical clustering to identify explanatory variables and perform dimensionality reduction [99], it was shown that monocyte recovery correlated with microaerophilic Lactobacillus spp. in stool. Supporting this association, is empirical evidence that the pili of Lactobacillus spp. known to thrive in microaerophilic conditions in the gut, such as Lactobacillus rhamnosus GG, stimulate monocytes via TLR2 [100]. Using another iterative technique that facilitates the integration of categorical and continuous clinical data [75], canonical correspondence analysis (CCpnA), Ingham et al. derived associations between the GM, immune recovery and GVHD post-allo-HSCT. CCpnA clustering was similar to that observed with sPLS, and revealed an association between severe GVHD, high monocyte counts and Lactobacillus spp. [7]. It would be interesting to further define M1/M2 polarization of macrophage recovery in this study, given that orally administered Lactobacillus spp. have been shown to drive M1 polarization in murine bone marrow derived macrophages [101] and M1 polarization has also been associated with GVHD in mice and humans [102,103]. Assessing adaptive immune responses, Ingham et al. also showed that early NK and B cell reconstitution were associated with less severe GVHD and a predominance of obligate anaerobes of the clostridial order Ruminococcacae and Lachnospiracae in the GM [7]. B cell expansion can be stimulated by superantigens from these species; however, the precise role of B cells in GVHD is more complex—early naïve B cell recovery may be associated with less severe GVHD, whilst continued B cell secretion of autoantibodies may drive chronic GVHD in the later months post-allo-HSCT [29]. Further studies assessing the precise immunophenotype and transcriptome of these cells may elucidate the exact immune subsets and potential mechanisms driving these associations in this pediatric cohort.
The mechanisms by which alterations in the GM impact immune reconstitution in humans is mostly speculative. Presence of microbial-derived short chain fatty acids, such as butyrate, in serum of patients have recently been shown to be inversely associated with chronic GVHD; however, the effect of butyrate on Treg reconstitution in thus study was not shown [104]. We and others have shown bacteria capable of producing riboflavin metabolites are associated with MAIT cell recovery and decreased severity of GVHD, supporting existing empirical observations in mouse models [34,59,105]. Using PICRUSt—a bioinformatic tool to predict metabolic pathways present in organisms sequenced by 16Sseq [106]—Konuma et al. showed that organisms that would be predicted to express the riboflavin metabolizing genes, ribA and ribB, were associated with MAIT cell recovery. However, without gene-level evidence of involvement using SMGS, such functional effect is unable to be confirmed.
CONCLUSIONS AND FUTURE DIRECTIONS
There is a large and growing body of evidence linking GM composition with normal immune development. Similar associations have been reported in interactions between the GM and post-allo-HSCT immune reconstitution and outcomes, yet gaps remain. Identification of immune subsets that are affected by distinct microbial species may guide directed FMT, pre-biotic and post-biotic therapies post-allo-HSCT. In addition to new technologies such as SMGS and iterative statistical methods that take advantage of increasing computational power will be able to identify associations in large, multi-table microbial and clinical datasets in relatively small groups of patients without being subject to increased identification of false positives [75]. Utilizing more robust identification of species- and gene-specific associations with immunophenotypes, transcriptomes and clinical outcomes and correlating these data with findings in normal immune development may lead to directed manipulations of the GM and/or metabolic pathways that will improve immune reconstitution and outcomes following allo-HSCT.
ACKNOWLEDGEMENTS
Thank you to Dr Samuel Minot for valuable discussions regarding microbial sequencing and Dr Stefanie Kalfas for manuscript editing.
FUNDING
This work was funded by the National Institutes of Health Research Project Grant (R01): The colonic microbiota and immunity after hematopoietic stem cell transplantation (R01 HL132350).
Footnotes
CONFLICTS OF INTEREST
The authors declare no relevant conflicts of interest.
REFERENCES
- 1.Gooley TA, Chien JW, Pergam SA, Hingorani S, Sorror ML, Boeckh M, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363(22):2091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carreras E, Dufour C, Mohty M, Kröger N, editors. The EBMT Handbook Hematopoietic Stem Cell Transplantation and Cellular Therapies. Cham (CH): Springer; 2019. [PubMed] [Google Scholar]
- 3.Dekker L, de Koning C, Lindemans C, Nierkens S. Reconstitution of T Cell Subsets Following Allogeneic Hematopoietic Cell Transplantation. Cancers. 2020. July 20;12(7):1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyiadzis M, Arora M, Klein JP, Hassebroek A, Hemmer M, Urbano-Ispizua A, et al. Impact of Chronic Graft-versus-Host Disease on Late Relapse and Survival on 7,489 Patients after Myeloablative Allogeneic Hematopoietic Cell Transplantation for Leukemia. Clin Cancer Res. 2015;21(9):2020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronan V, Yeasin R, Claud EC. Childhood Development and the Microbiome: The Intestinal Microbiota in Maintenance of Health and Development of Disease during Childhood Development. Gastroenterology. 2020;S0016-5085(20)35526–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staffas A, Burgos da Silva M, Slingerland AE, Lazrak A, Bare CJ, Holman CD, et al. Nutritional Support from the Intestinal Microbiota Improves Hematopoietic Reconstitution after Bone Marrow Transplantation in Mice. Cell Host Microbe. 2018;23(4):447–57. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingham AC, Kielsen K, Cilieborg MS, Lund O, Holmes S, Aarestrup FM, et al. Specific gut microbiome members are associated with distinct immune markers in pediatric allogeneic hematopoietic stem cell transplantation. Microbiome. 2019;7(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, et al. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588(7837):303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogonek J, Kralj Juric M, Ghimire S, Varanasi PR, Holler E, Greinix H, et al. Immune Reconstitution after Allogeneic Hematopoietic Stem Cell Transplantation. Front Immunol. 2016. November 17;7:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rezvani AR, Storer BE, Guthrie KA, Schoch HG, Maloney DG, Sandmaier BM, et al. Impact of donor age on outcome after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2015;21(1):105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun YQ, Wang Y, Zhang XH, Xu LP, Liu KY, Yan CH, et al. Virus reactivation and low dose of CD34+ cell, rather than haploidentical transplantation, were associated with secondary poor graft function within the first 100 days after allogeneic stem cell transplantation. Ann Hematol. 2019;98(8):1877–83. [DOI] [PubMed] [Google Scholar]
- 12.Singh V, Jang H, Kim S, Ayash L, Alavi A, Ratanatharathorn V, et al. G-CSF use post peripheral blood stem cell transplant is associated with faster neutrophil engraftment, shorter hospital stay and increased incidence of chronic GVHD. Leuk Lymphoma. 2020:1–8. [DOI] [PubMed] [Google Scholar]
- 13.Minculescu L, Fischer-Nielsen A, Haastrup E, Ryder LP, Andersen NS, Schjoedt I, et al. Improved Relapse-Free Survival in Patients With High Natural Killer Cell Doses in Grafts and During Early Immune Reconstitution After Allogeneic Stem Cell Transplantation. Front Immunol. 2020;11:1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ando T, Tachibana T, Tanaka M, Suzuki T, Ishiyama Y, Koyama S, et al. Impact of graft sources on immune reconstitution and survival outcomes following allogeneic stem cell transplantation. Blood Adv. 2020;4(2):408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeCook LJ, Thoma M, Huneke T, Johnson ND, Wiegand RA, Patnaik MM, et al. Impact of lymphocyte and monocyte recovery on the outcomes of allogeneic hematopoietic SCT with fludarabine and melphalan conditioning. Bone Marrow Transplant. 2013;48(5):708–14. [DOI] [PubMed] [Google Scholar]
- 16.Thoma MD, Huneke TJ, DeCook LJ, Johnson ND, Wiegand RA, Litzow MR, et al. Peripheral Blood Lymphocyte and Monocyte Recovery and Survival in Acute Leukemia Postmyeloablative Allogeneic Hematopoietic Stem Cell Transplant. Biol Blood Marrow Transplant. 2012;18(4):600–7. [DOI] [PubMed] [Google Scholar]
- 17.Dhakal B, Brazauskas R, Lara CA, Hari P, Pasquini M, D’Souza A. Monocyte recovery at day 100 is associated with improved survival in multiple myeloma patients who undergo allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. 2016;51(2):297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TW, Park SS, Lim JY, Min GJ, Park S, Jeon YW, et al. Predictive Role of Circulating Immune Cell Subtypes Early after Allogeneic Hematopoietic Stem Cell Transplantation in Patients with Acute Leukemia. Int J Stem Cells. 2018;12(1):73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varanasi PR, Ogonek J, Luther S, Dammann E, Stadler M, Ganser A, et al. Cytomegalovirus-specific CD8+ T-cells are associated with a reduced incidence of early relapse after allogeneic stem cell transplantation. PLoS One. 2019;14(3):e0213739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkman R, Cohen G, Carter SL, Weinberg KI, Masinsin B, Guinan E, et al. Successful immune reconstitution decreases leukemic relapse and improves survival in recipients of unrelated cord blood transplantation. Biol Blood Marrow Transplant. 2006;12(9):919–27. [DOI] [PubMed] [Google Scholar]
- 21.Kim DH, Sohn SK, Won DI, Lee NY, Suh JS, Lee KB. Rapid helper T-cell recovery above 200 × 10 6/l at 3 months correlates to successful transplant outcomes after allogeneic stem cell transplantation. Bone Marrow Transplant. 2006;37(12):1119–28. [DOI] [PubMed] [Google Scholar]
- 22.Waller EK, Logan BR, Fei M, Lee SJ, Confer D, Howard A, et al. Kinetics of immune cell reconstitution predict survival in allogeneic bone marrow and G-CSF-mobilized stem cell transplantation. Blood Adv. 2019;3(15):2250–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alho AC, Kim HT, Chammas MJ, Reynolds CG, Matos TR, Forcade E, et al. Unbalanced recovery of regulatory and effector T cells after allogeneic stem cell transplantation contributes to chronic GVHD. Blood. 2016;127(5):646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamae H, Fujii K, Nanno S, Okamura H, Nakane T, Koh H, et al. A prospective observational study of immune reconstitution following transplantation with post-transplant reduced-dose cyclophosphamide from HLA-haploidentical donors. Transpl Int. 2019;32(12):1322–32. [DOI] [PubMed] [Google Scholar]
- 25.Kawaguchi K, Umeda K, Hiejima E, Iwai A, Mikami M, Nodomi S, et al. Influence of post-transplant mucosal-associated invariant T cell recovery on the development of acute graft-versus-host disease in allogeneic bone marrow transplantation. Int J Hematol. 2018;108(1):66–75. [DOI] [PubMed] [Google Scholar]
- 26.Perko R, Kang G, Sunkara A, Leung W, Thomas PG, Dallas MH. Gamma delta T cell reconstitution is associated with fewer infections and improved event-free survival after hematopoietic stem cell transplantation for pediatric leukemia. Biol Blood Marrow Transplant. 2015;21(1):130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bian Z, Xu LP, Fu Q, Huo M, Liu L, Zhao X, et al. Homeostatic γδ T Cell Contents Are Preserved by Granulocyte Colony-Stimulating Factor Priming and Correlate with the Early Recovery of γδ T Cell Subsets after Haploidentical Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2018;24(2):252–9. [DOI] [PubMed] [Google Scholar]
- 28.Arruda LCM, Gaballa A, Uhlin M. Impact of γδ T cells on clinical outcome of hematopoietic stem cell transplantation: systematic review and meta-analysis. Blood Adv. 2019;3(21):3436–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Maas NG, Berghuis D, van der Burg M, Lankester AC. B Cell Reconstitution and Influencing Factors After Hematopoietic Stem Cell Transplantation in Children. Front Immunol. 2019;10:782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagayama T, Fujiwara SI, Ikeda T, Kawaguchi SI, Toda Y, Ito S, et al. Steep neutrophil recovery following unrelated bone marrow transplantation is a major risk factor for the development of acute graft-vs-host disease-a retrospective study. Transpl Int. 2020. September 16. doi: 10.1111/tri.13741 [DOI] [PubMed] [Google Scholar]
- 31.Turcotte FM, Cao Q, Cooley SA, Curtsinger J, Holtan SG, Fuo X, et al. Monocyte Subpopulation Recovery as Predictors of Hematopoietic Cell Transplantation Outcomes. Biol Blood Marrow Transplant. 2019;25(5):883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohty M, Blaise D, Faucher C, Bardou VJ, Gastaut JA, Viens P, et al. Impact of plasmacytoid dendritic cells on outcome after reduced-intensity conditioning allogeneic stem cell transplantation. Leukemia. 2005;19(1):1–6. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe N, Narita M, Furukawa T, Nakamura T, Yamahira A, Masuko M, et al. Kinetics of pDCs, mDCs, γδT cells and regulatory T cells in association with graft versus host disease after hematopoietic stem cell transplantation. Int J Fab Hematol. 2011;33(4):378–90. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharyya A, Hanafi FA, Sheih A, Golob JF, Srinivasan S, Boeckh MJ, et al. Graft-Derived Reconstitution of Mucosal-Associated Invariant T Cells after Allogeneic Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant. 2018;24(2):242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez FD, Holt PG. Role of microbial burden in aetiology of allergy and asthma. Lancet. 1999;354(Suppl 2):Sii12–5. [DOI] [PubMed] [Google Scholar]
- 36.von Mutius E, Martinez FD, Fritzsch C, Nicolai T, Roell G, Thiemann HH. Prevalence of asthma and atopy in two areas of West and East Germany. Am J Respir Crit Care Med. 1994;149(2 Pt 1):358–64. [DOI] [PubMed] [Google Scholar]
- 37.Dobson GP, Letson HE, Biros E, Morris J. Specific pathogen-free (SPF) animal status as a variable in biomedical research: Have we come full circle? EBioMedicine. 2019;41:42–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DF. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–18. [DOI] [PubMed] [Google Scholar]
- 39.Ennamorati M, Vasudevan C, Clerkin K, Halvorsen S, Verma S, Ibrahim S, et al. Intestinal microbes influence development of thymic lymphocytes in early life. Proc Natl Acad Sci USA. 2020;117(5):2570–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakrishna C, Kujawski M, Chu H, Fi F, Mazmanian SK, Cantin EM. Bacteroides fragilis polysaccharide A induces IF-10 secreting B and T cells that prevent viral encephalitis. Nat Commun. 2019;10(1):2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163(2):367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Britton GJ, Contijoch EJ, Spindler MP, Aggarwala V, Dogan B, Bongers G, et al. Defined microbiota transplant restores Th17/RORγt(+) regulatory T cell balance in mice colonized with inflammatory bowel disease microbiotas. Proc Natl Acad Sci U S A. 2020;117(35):21536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giesbrecht K, Förmer S, Sähr A, Heeg K, Hildebrand D. Streptococcal Pyrogenic Exotoxin A-Stimulated Monocytes Mediate Regulatory T-Cell Accumulation through PD-L1 and Kynurenine. Int J Mol Sci. 2019;20(16):3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20(5):524–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang TWH, Chen HC, Chen CY, Yen CYT, Lin CJ, Prajnamitra RP, et al. Loss of Gut Microbiota Alters Immune System Composition and Cripples Postinfarction Cardiac Repair. Circulation. 2019;139(5):647–59. [DOI] [PubMed] [Google Scholar]
- 46.Tsugawa H, Kabe Y, Kanai A, Sugiura Y, Hida S, Taniguchi S, et al. Short-chain fatty acids bind to apoptosis-associated speck-like protein to activate inflammasome complex to prevent Salmonella infection. PLoS Biol. 2020;18(9):e3000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Negi S, Pahari S, Bashir H, Agrewala JN. Gut Microbiota Regulates Mincle Mediated Activation of Lung Dendritic Cells to Protect Against Mycobacterium tuberculosis. Front Immunol. 2019;10:1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fink LN, Zeuthen LH, Christensen HR, Morandi B, Frøkiaer H, Ferlazzo G. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated NK cell responses. Int Immunol. 2007;19(12):1319–27. [DOI] [PubMed] [Google Scholar]
- 49.Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143(2):418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019;366(6464):494–9. [DOI] [PubMed] [Google Scholar]
- 51.Eckle SB, Birkinshaw RW, Kostenko L, Corbett AJ, McWilliam HE, Reantragoon R, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med. 2014;211(8):1585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8(+) T Cells. Immunity. 2019;51(2):285–97. e5. [DOI] [PubMed] [Google Scholar]
- 53.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328(5986):1705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Limenitakis JP, Greiff V, Yilmaz B, Scharen O, Urbaniak C, et al. Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature. 2020;584(7820):274–8. [DOI] [PubMed] [Google Scholar]
- 56.Nagashima K, Sawa S, Nitta T, Tsutsumi M, Okamura T, Penninger JM, et al. Identification of subepithelial mesenchymal cells that induce IgA and diversify gut microbiota. Nat Immunol. 2017;18(6):675–82. [DOI] [PubMed] [Google Scholar]
- 57.Lindner C, Thomsen I, Wahl B, Ugur M, Sethi MK, Friedrichsen M, et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol. 2015;16(8):880–8. [DOI] [PubMed] [Google Scholar]
- 58.Lindner C, Wahl B, Fohse L, Suerbaum S, Macpherson AJ, Prinz I, et al. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med. 2012;209(2):365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varelias A, Bunting MD, Ormerod KL, Koyama M, Olver SD, Straube J, et al. Recipient mucosal-associated invariant T cells control GVHD within the colon. J Clin Invest. 2018;128(5):1919–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581(7809):475–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niu X, Daniel S, Kumar D, Ding EY, Savani RC, Koh AY, et al. Transient neonatal antibiotic exposure increases susceptibility to late-onset sepsis driven by microbiota-dependent suppression of type 3 innate lymphoid cells. Sci Rep. 2020;10(1):12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J Clin Oncol. 2017;35(15):1650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masetti R, Zama D, Leardini D, Muratore E, Turroni S, Prete A, et al. The gut microbiome in pediatric patients undergoing allogeneic hematopoietic stem cell transplantation. Pediatr Blood Cancer. 2020. December;67(12):e28711. [DOI] [PubMed] [Google Scholar]
- 64.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20(5):640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209(5):903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gavriilaki M, Sakellari I, Anagnostopoulos A, Gavriilaki E. The Impact of Antibiotic-Mediated Modification of the Intestinal Microbiome on Outcomes of Allogeneic Hematopoietic Cell Transplantation: Systematic Review and Meta-Analysis. Biol Blood Marrow Transplant. 2020;26(9):1738–46. [DOI] [PubMed] [Google Scholar]
- 67.Fischer JC, Bscheider M, Eisenkolb G, Lin CC, Wintges A, Otten V, et al. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci Transl Med. 2017;9(386). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eriguchi Y, Nakamura K, Hashimoto D, Shimoda S, Shimono N, Akashi K, et al. Decreased secretion of Paneth cell alpha-defensins in graft-versus-host disease. Transpl Infect Dis. 2015;17(5):702–6. [DOI] [PubMed] [Google Scholar]
- 69.Andersen S, Staudacher H, Weber N, Kennedy G, Varelias A, Banks M, et al. Pilot study investigating the effect of enteral and parenteral nutrition on the gastrointestinal microbiome post-allogeneic transplantation. Br J Haematol. 2020;188(4):570–81. [DOI] [PubMed] [Google Scholar]
- 70.Nguyen NP, Warnow T, Pop M, White B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microbiomes. 2016;2:16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med. 2020;382(9):822–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith NW, Shorten PR, Altermann E, Roy NC, McNabb WC. The Classification and Evolution of Bacterial Cross-Feeding. Front Ecol Evol. 2019;7:153. [Google Scholar]
- 73.Minot SS, Willis AD. Clustering co-abundant genes identifies components of the gut microbiome that are reproducibly associated with colorectal cancer and inflammatory bowel disease. Microbiome. 2019;7(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Willis AD, Minot SS. Strategies to Facilitate Translational Advances from Microbiome Surveys. Trends Microbiol. 2020;28(5):329–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sankaran K, Holmes SP. Multitable Methods for Microbiome Data Integration. Frontiers in Genetics. 2019;10(627). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin BD, Witten D, Willis AD. MODELING MICROBIAL ABUNDANCES AND DYSBIOSIS WITH BETA-BINOMIAL REGRESSION. Ann Appl Stat. 2020;14(1):94–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8(339):339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwab L, Goroncy L, Palaniyandi S, Gautam S, Triantafyllopoulou A, Mocsai A, et al. Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance graft-versus-host disease via tissue damage. Nat Med. 2014;20(6):648–54. [DOI] [PubMed] [Google Scholar]
- 79.Bowerman KL, Varelias A, Lachner N, Kuns RD, Hill GR, Hugenholtz P. Continuous pre- and post-transplant exposure to a disease-associated gut microbiome promotes hyper-acute graft-versus-host disease in wild-type mice. Gut Microbes. 2020;11(4):754–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koyama M, Mukhopadhyay P, Schuster IS, Henden AS, Hülsdünker J, Varelias A, et al. MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft-versus-Host Disease and Is Influenced by the Microbiota. Immunity. 2019;51(5):885–98. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, et al. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science. 2019;366(6469):1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129(6):729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwamura C, Bouladoux N, Belkaid Y, Sher A, Jankovic D. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 2017;129(2):171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mancini N, Greco R, Pasciuta R, Barbanti MC, Pini G, Morrow OB, et al. Enteric Microbiome Markers as Early Predictors of Clinical Outcome in Allogeneic Hematopoietic Stem Cell Transplant: Results of a Prospective Study in Adult Patients. Open Forum Infect Dis. 2017;4(4):ofx215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Payen M, Nicolis I, Robin M, Michonneau D, Delannoye J, Mayeur C, et al. Functional and phylogenetic alterations in gut microbiome are linked to graft-versus-host disease severity. Blood Adv. 2020;4(9):1824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Golob JL, Pergam SA, Srinivasan S, Fiedler TL, Liu C, Garcia K, et al. Stool Microbiota at Neutrophil Recovery Is Predictive for Severe Acute Graft vs Host Disease After Hematopoietic Cell Transplantation. Clin Infect Dis. 2017;65(12):1984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu C, Frank DN, Horch M, Chau S, Ir D, Horch EA, et al. Associations between acute gastrointestinal GvHD and the baseline gut microbiota of allogeneic hematopoietic stem cell transplant recipients and donors. Bone Marrow Transplant. 2017;52(12):1643–50. [DOI] [PubMed] [Google Scholar]
- 89.Han L, Zhao K, Li Y, Han H, Zhou L, Ma P, et al. A gut microbiota score predicting acute graft-versus-host disease following myeloablative allogeneic hematopoietic stem cell transplantation. Am J Transplant. 2020;20(4):1014–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doki N, Suyama M, Sasajima S, Ota J, Igarashi A, Mimura I, et al. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann Hematol. 2017;96(9):1517–23. [DOI] [PubMed] [Google Scholar]
- 91.Weber D, Jenq RR, Peled JU, Taur Y, Hiergeist A, Koestler J, et al. Microbiota Disruption Induced by Early Use of Broad-Spectrum Antibiotics Is an Independent Risk Factor of Outcome after Allogeneic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2017;23(5):845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tamburini FB, Andermann TM, Tkachenko E, Senchyna F, Banaei N, Bhatt AS. Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat Med. 2018;24(12):1809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kakihana K, Fujioka Y, Suda W, Najima Y, Kuwata G, Sasajima S, et al. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood. 2016;128(16):2083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshifuji K, Inamoto K, Kiridoshi Y, Takeshita K, Sasajima S, Shiraishi Y, et al. Prebiotics protect against acute graft-versus-host disease and preserve the gut microbiota in stem cell transplantation. Blood Adv. 2020;4(19):4607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.DeFilipp Z, Peled JU, Li S, Mahabamunuge J, Dagher Z, Slingerland AE, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv. 2018;2(7):745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Lier YF, Davids M, Haverkate NJE, de Groot PF, Donker ML, Meijer E, et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci Transl Med. 2020;12(556). [DOI] [PubMed] [Google Scholar]
- 97.Simms-Waldrip TR, Sunkersett G, Coughlin LA, Savani MR, Arana C, Kim J, et al. Antibiotic-Induced Depletion of Anti-inflammatory Clostridia Is Associated with the Development of Graft-versus-Host Disease in Pediatric Stem Cell Transplantation Patients. Biol Blood Marrow Transplant. 2017;23(5):820–9. [DOI] [PubMed] [Google Scholar]
- 98.Biagi E, Zama D, Rampelli S, Turroni S, Brigidi P, Consolandi C, et al. Early gut microbiota signature of aGvHD in children given allogeneic hematopoietic cell transplantation for hematological disorders. BMC Med Genomics. 2019;12(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chun H, Keleş S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J R Stat Soc Series B Stat Methodol. 2010;72(l):3–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.von Ossowski I, Pietilä TE, Rintahaka J, Nummenmaa E, Mäkinen VM, Reunanen J, et al. Using recombinant Lactococci as an approach to dissect the immunomodulating capacity of surface piliation in probiotic Lactobacillus rhamnosus GG. PLoS One. 2013;8(5):e64416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang B, Wu Y, Liu R, Xu H, Mei X, Shang Q, et al. Lactobacillus rhamnosus GG promotes Ml polarization in murine bone marrow-derived macrophages by activating TLR2/MyD88/MAPK signaling pathway. Anim Sci J. 2020;91(1):e13439. [DOI] [PubMed] [Google Scholar]
- 102.Seno K, Yasunaga M, Kajiya H, Izaki-Hagio K, Morita H, Yoneda M, et al. Dynamics of M1 macrophages in oral mucosal lesions during the development of acute graft-versus-host disease in rats. Clin Exp Immunol. 2017;190(3):315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wen Q, Kong Y, Zhao HY, Zhang YY, Han TT, Wang Y, et al. G-CSF-induced macrophage polarization and mobilization may prevent acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2019;54(9):1419–33. [DOI] [PubMed] [Google Scholar]
- 104.Markey KA, Schluter J, Gomes ALC, Littmann ER, Pickard AJ, Taylor BP, et al. The microbe-derived short-chain fatty acids butyrate and propionate are associated with protection from chronic GVHD. Blood. 2020;136(1):130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Konuma T, Kohara C, Watanabe E, Takahashi S, Ozawa G, Suzuki K, et al. Reconstitution of Circulating Mucosal-Associated Invariant T Cells after Allogeneic Hematopoietic Cell Transplantation: Its Association with the Riboflavin Synthetic Pathway of Gut Microbiota in Cord Blood Transplant Recipients. J Immunol. 2020;204(6):1462–73. [DOI] [PubMed] [Google Scholar]
- 106.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]


