Fig. 3. Mass spectrometric analysis of S-nitrosylated proteins in rat AVICs exposed to NO donor demonstrates an enrichment of ubiquitin-specific processing proteases and identifies USP9X.
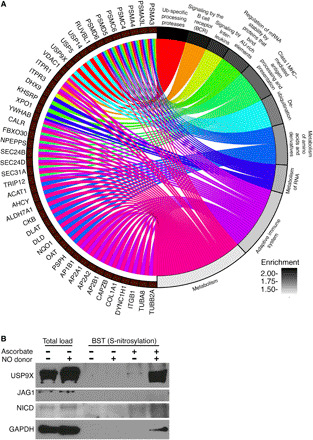
(A) Chord plot illustrates overrepresentation of NO-enriched S-nitrosylated proteins. Top 10 Gene Ontology pathways (GO Slim) and gene sets are shown. (B) Immunoblot shows S-nitrosylation of USP9X in the presence of NO donor, where negative ascorbate serves as a technical negative control. NICD and JAG1 immunoblots with ascorbate serve as biological negative controls. GAPDH, a known S-nitrosylated protein, serves as a biological positive control. Total load represents the initial cell lysates used to purify S-nitrosylated proteins. AU-rich element, adenylate/uridylate-rich elements.
