Figure 2.
Replication of pMH is a two-step mutagenic process
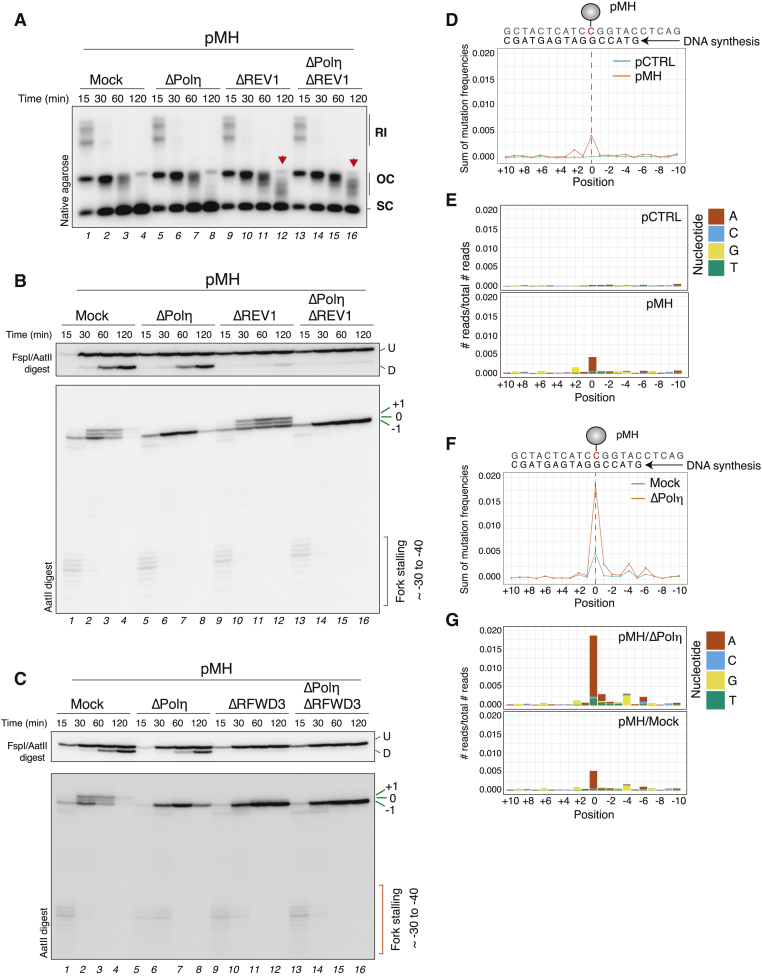
(A) pMH was replicated in mock-, Polη-, REV1-, or Polη- and REV1-depleted extracts. Samples were analyzed as in Figure 1C.
(B) Samples from (A) were digested and analyzed as in Figure 1E. U, undamaged strand; D, damaged strand.
(C) pMH was replicated in mock-, Polη-, RFWD3-, or Polη- and RFWD3-depleted extracts. Samples were analyzed as in Figure 1E.
(D) Quantification of mutation frequencies measured after replication of pCTRL or pMH. Replication samples were amplified by PCR and analyzed by next-generation sequencing (see STAR methods). The 0 position corresponds to the location of the protein adduct. One of three independent experiments is shown.
(E) Distribution of nucleotide misincorporation from the data generated in (D).
(F) pMH was replicated in either mock- or Polη-depleted extracts, and samples were amplified and analyzed as in (D). One of three independent experiments is shown.
(G) Distribution of nucleotide misincorporation from the data generated in (F).