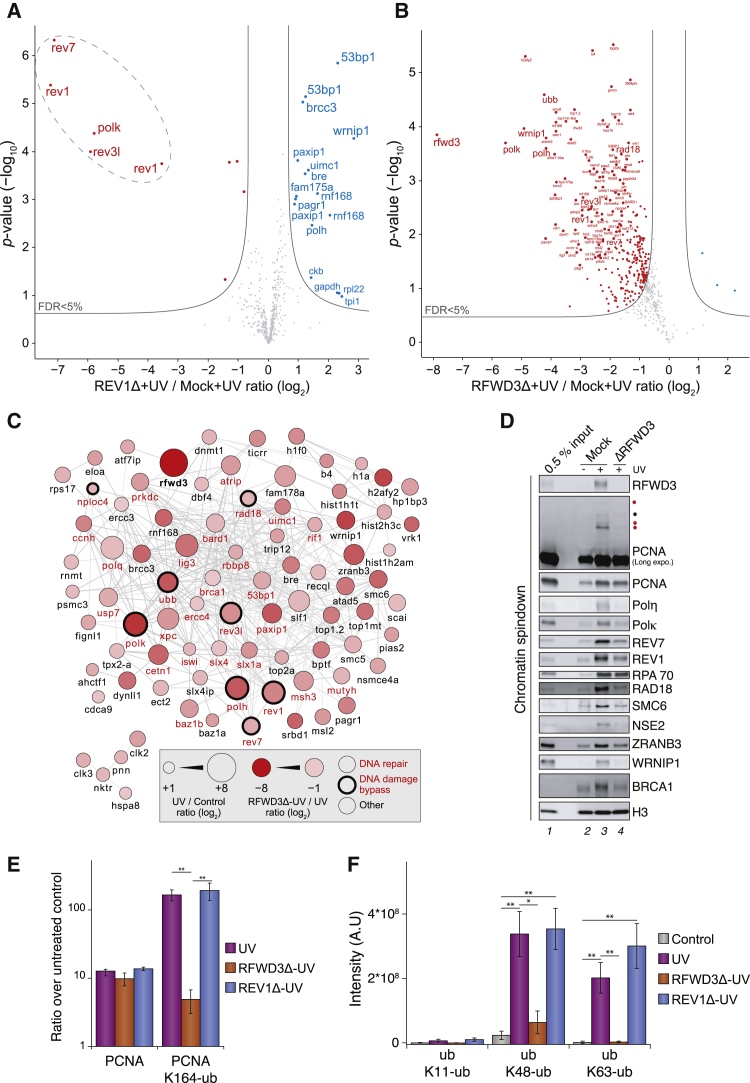
Figure 6.
RFWD3 regulates ubiquitin levels and protein recruitment to UV damaged chromatin
(A) MS analysis of protein recruitment to UV-treated sperm chromatin in mock- or REV1-depleted extracts. The volcano plot shows the difference in abundance of proteins between the two sample conditions (x axis), plotted against the p value resulting from two-tailed Student’s t testing (y axis). Proteins significantly down- or upregulated (false discovery rate [FDR] < 5%) in REV1-depleted reactions are represented in red or blue, respectively. n = 4 biochemical replicates. FDR < 5% corresponds to permutation-based, FDR-adjusted q < 0.05. Different isoforms of the same protein can be detected (e.g., REV1).
(B) Same experiment as in (A) but comparing mock- to RFWD3-depleted extracts. Small red dots, 1% < FDR < 5%; large red dots, FDR < 1%.
(C) STRING network of proteins highly significantly upregulated (FDR < 1%) on UV-treated sperm chromatin compared with mock-treated chromatin and highly significantly downregulated (FDR < 1%) on UV-treated sperm chromatin with RFWD3 depletion compared with mock depletion.
(D) An independent experiment was analyzed by immunoblot using the indicated antibodies.
(E) Average PCNA and PCNA K164-ubiquitin abundance on sperm chromatin identified by tandem mass spectrometry (MS/MS), quantified in a label-free manner, and plotted as a ratio over untreated control. n = 4 biochemical replicates. Error bars represent SEM. ∗∗p < 0.01, via two-tailed Student’s t testing.
(F) Quantification of ubiquitin linkages on sperm chromatin directly identified by MS/MS via diglycine-modified lysine residues in the corresponding peptide sequences and quantified in a label-free manner. n = 4 biochemical replicates. Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01, via two-tailed Student’s t testing.