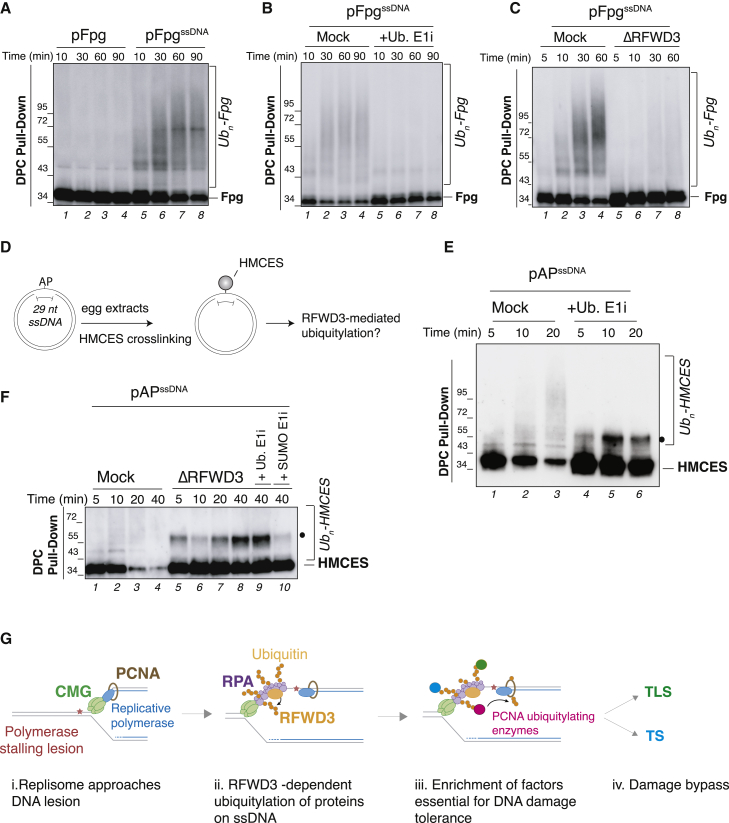
Figure 7.
RFWD3 simulates ubiquitylation of proteins on ssDNA
(A) Fpg bacterial glycosylase was crosslinked to either double-stranded (pFpg) or single-stranded DNA (pFpgssDNA) and added to SPRTN-depleted non-licensing egg extracts. DPC pull-down under stringent conditions was performed at the indicated time points, and samples were blotted against crosslinked Fpg. Slow mobility bands represent ubiquitylated Fpg species (see B).
(B) pFpgssDNA was incubated in SPRTN-depleted non-licensing extracts, and ubiquitin E1 inhibitor was added where indicated. Plasmids were recovered, and samples were blotted against Fpg as in (A).
(C) pFpgssDNA was incubated in mock- or RFWD3-depleted non-licensing extracts (also depleted of SPRTN) for the indicated time points and samples processed as in (A).
(D) Generation of an AP site on ssDNA (pAPssDNA) to induce HMCES crosslinking.
(E) pAPssDNA was incubated in SPRTN-depleted non-licensing extracts, and ubiquitin E1 inhibitor was added where indicated. Plasmids were recovered, and proteins were blotted against HMCES. The black dot indicates sumoylated HMCES (see F).
(F) pAPssDNA was incubated in mock- or RFWD3-depleted non-licensing extracts (depleted of SPRTN), and ubiquitin E1 inhibitor or SUMO E1 inhibitor was added where indicated. Plasmids were recovered and analyzed as in (D).
(G) Model illustrating the role of RFWD3 in gap-filling DNA synthesis (see Discussion).