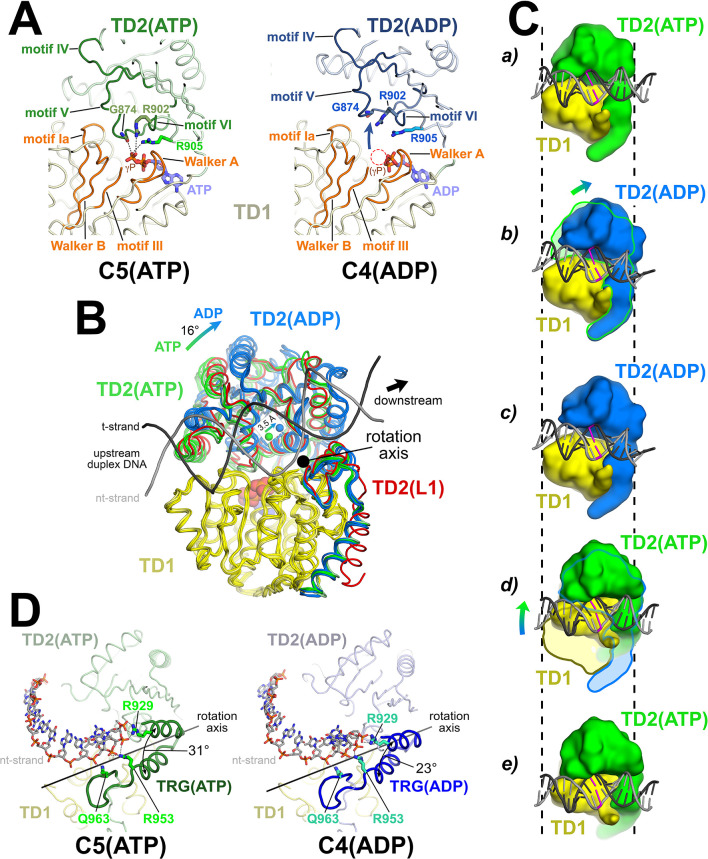
Figure 3. The Mfd translocation module and the DNA translocation mechanism.
(A) Conformational changes of the Mfd translocation module induced by ATP hydrolysis and Pi release. The structural environments of the ATP [left; C5(ATP)] or ADP [right; C4(ADP)] binding sites are shown. The protein is shown as a backbone worm; TD1 is colored pale yellow but the SF2 ATPase motifs of TD1 (Walker A, motif Ia, Walker B, motif III) are colored orange; TD2(ATP) or (ADP) are colored pale green or light blue, respectively, but the SF2 ATPase motifs of TD2 (motifs IV, V, and VI) are colored dark green or dark blue. The nucleotide is shown in stick format with blue carbon atoms. The side chain or backbone atoms of three key residues, G874 (motif V), R902 (motif VI), and R905 (just beyond motif VI), are also shown. The backbone carbonyl of G874 and the side chains of R902 and R905 form polar interactions with the ATP γ-phosphate (denoted as gray dashed lines). In the ADP structure, these interactions are lost due to the missing γ-phosphate (denoted by the dashed red circle), causing TD2 to swing away from TD1 (denoted by the thick arrow). Here, 'polar contacts' include both hydrogen bonds (≤3.5 Å) and ionic interactions. Ionic interactions can include both close range interactions (where the hydration shells of the two oppositely charged moieties are displaced) that are typically called salt-bridges (≤3.5 Å), but can also include longer range interactions where the two oppositely charged moieties remain hydrated but their Coulombic interaction is favorable (Kumar and Nussinov, 2002; Xu et al., 1997). These longer-range favorable interactions are significant and extend to well beyond 4.5 Å distance (Yu et al., 2019). (B) The translocation modules of all seven Mfd-elongation complex (EC) structures were superimposed by alignment of TD1 (colored yellow) α-carbons. The resulting positions of TD2 clustered into two groups, those with ATP (TD2 colored green) or ADP (TD2 colored blue). TD2 of L1 is shown in red and clusters with the ATP-bound structures. The relative disposition of the upstream duplex DNA is also shown (gray phosphate backbone worms). TD2(ATP) and TD2(ADP) are related by an ~16° rotation (denoted by the thick arrow, upper left) about an axis roughly perpendicular to the DNA helical axis (denoted by the black dot), resulting in a 3.5 Å shift of the TD2 center of mass roughly parallel to the DNA helical axis [center of mass positions for TD2(ATP) and TD2(ADP) denoted by the green and blue spheres, respectively], corresponding to one base pair rise of B-form DNA. The 3.5 Å shift of the TD2 center of mass is not sensitive to which structures are used to perform the calculation. (C) Inchworm model for duplex DNA translocation. Duplex DNA is shown as a cartoon (for reference, a central base pair is colored magenta). TD1 is colored yellow, while TD2 is colored green (ATP) or blue (ADP). In (a), both TD1 and TD2(ATP) interact with the duplex DNA (the initial positions of TD1 and TD2 on the DNA are denoted by the vertical dashed reference lines). Upon ATP hydrolysis and Pi release, TD2(ADP) rotates away from TD1 (b) and interacts with the DNA one base pair downstream (to the right, c). With the release of ADP, ATP binding induces TD1 to rotate toward TD2 (d). In (e), TD1 and TD2(ATP) both interact with the duplex DNA but one base pair to the right. Also see Video 1. (D) Conformational changes of the TRG motif. Protein is shown as a backbone worm; TD1 is colored pale yellow; TD2(ATP) or (ADP) are colored pale green or light blue, respectively, but the TRG motifs are colored dark green or dark blue. The nt-strand of the upstream duplex DNA is shown in stick format (the t-strand of the DNA is not shown for clarity). Three key TRG motif residues interact with the nt-strand DNA backbone, R929, R953, and Q963 (side chains shown, polar interactions with the DNA denoted by the gray dashed lines). The rotation axis of the TD2(ATP) → TD2(ADP) conformational change passes directly through the TRG motif helical hairpin linker, which serves as the hinge. Opening of TD2(ADP) causes the TRG helical hairpin to pinch closed nearly 10 Å.