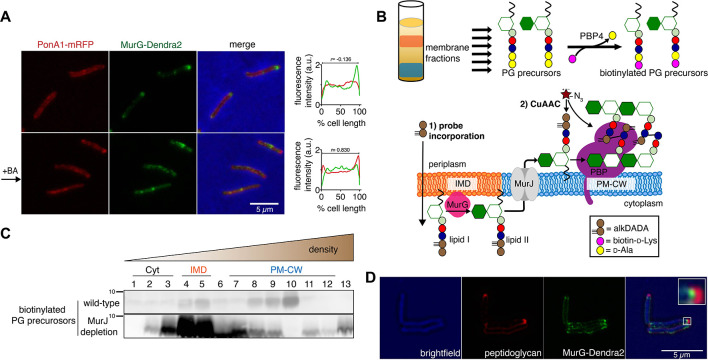
Figure 2. Lipid II is synthesized in the IMD and trafficked to the PM-CW.
(A) Left, conventional microscopy of M. smegmatis coexpressing PonA1-mRFP and MurG-Dendra2 treated +/- benzyl alcohol (BA). Right, fluorescence distribution of the fusion proteins. a.u., arbitrary units. r denotes the Pearson’s correlation value. 42>n>57. (B) Top, detection of lipid-linked peptidoglycan (PG) precursors from membrane fractions. Bottom, metabolic labeling of mycobacterial cell wall synthesis (García-Heredia et al., 2018). (C) PG precursors are labeled as in (B), top. The labeled precursors are in the IMD and PM-CW of wild-type M. smegmatis but accumulate in the IMD upon MurJ depletion (García-Heredia et al., 2018). While we do not yet understand the loss of signal from fraction 10, we note that there are precursors present but in low abundance (see Figure 2—figure supplement 3B). (D) M. smegmatis-expressing MurG-Dendra2 were incubated with alkDADA. Surface-exposed alkynes on fixed cells were detected by CuAAC (García-Heredia et al., 2018). Bacteria were imaged by SIM-E.