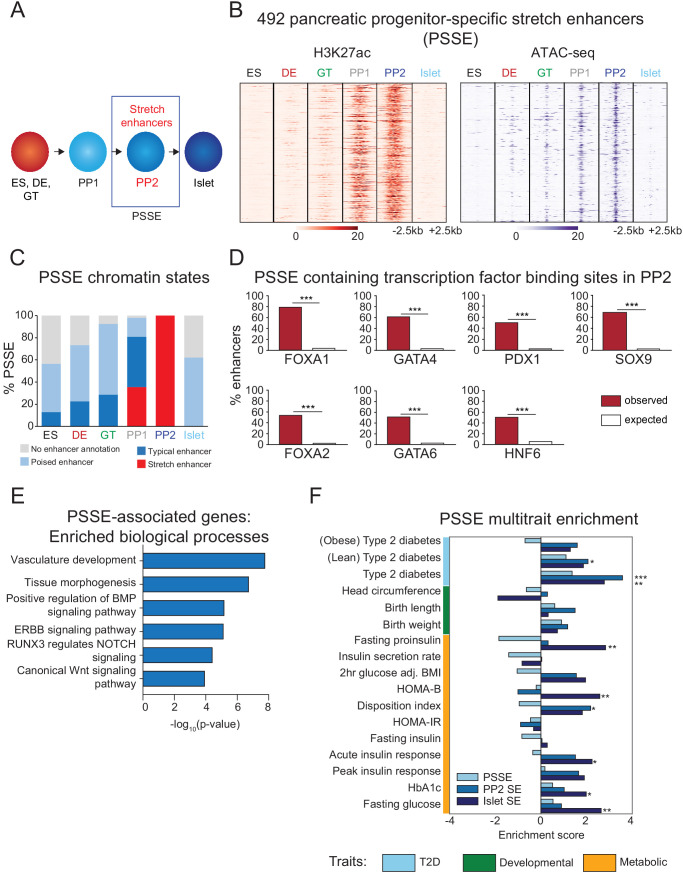
Figure 2. Candidate target genes of pancreatic progenitor-specific stretch enhancers regulate developmental processes.
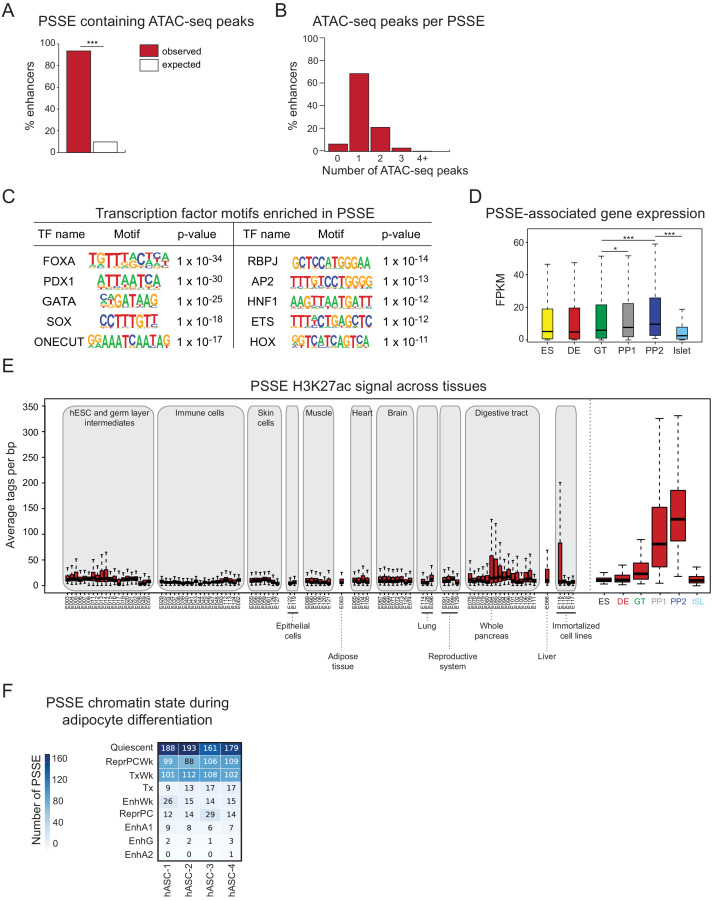
(A) Schematic illustrating identification of pancreatic progenitor-specific stretch enhancers (PSSE). (B) Heatmap showing density of H3K27ac ChIP-seq and ATAC-seq reads at PSSE, centered on overlapping H3K27ac and ATAC-seq peaks, respectively, and spanning 5 kb in ES, DE, GT, PP1, PP2, and islets. PSSE coordinates in Figure 2—source data 1. (C) Percentage of PSSE exhibiting indicated chromatin states at defined developmental stages and in islets. (D) Percentage of PSSE overlapping with at least one ChIP-seq peak at PP2 for the indicated transcription factors. Enrichment analysis comparing observed and expected overlap based on random genomic regions of the same size and located on the same chromosome averaged over 10,000 iterations (***p<1×10−4; permutation test). (E) Gene ontology analysis for nearest expressed genes (fragments per kilobase per million fragments mapped (FPKM) ≥1 at PP2) to the 492 PSSE. See also Figure 2—source data 2. (F) Enrichment (LD score regression coefficient z-scores) of T2D, developmental, and metabolic GWAS trait-associated variants at accessible chromatin sites in PSSE as compared with PP2 and islet stretch enhancers. Significant enrichment was identified within accessible chromatin at PP2 stretch enhancers for lean type 2 diabetes (Z = 2.06, *p=3.94 × 10−2), at PP2 stretch enhancers for type 2 diabetes (Z = 3.57, ***p=3.52 × 10−4), at islet stretch enhancers for type 2 diabetes (Z = 2.78, **p=5.46 × 10−3), at islet stretch enhancers for fasting proinsulin levels (Z = 2.83, **p=4.61 × 10−3), at islet stretch enhancers for HOMA-B (Z = 2.58, **p=9.85 × 10−3), at PP2 stretch enhancers for disposition index (Z = 2.18, *p=2.94 × 10−2), at islet stretch enhancers for acute insulin response (Z = 2.24, *p=2.51 × 10−2), at islet stretch enhancers for HbA1c (Z = 1.98, *p=4.72 × 10−2), and at islet stretch enhancers for fasting glucose levels (Z = 2.64, **p=8.31 × 10−3). See also Figure 2—source data 3 and Figure 2—figure supplement 1.