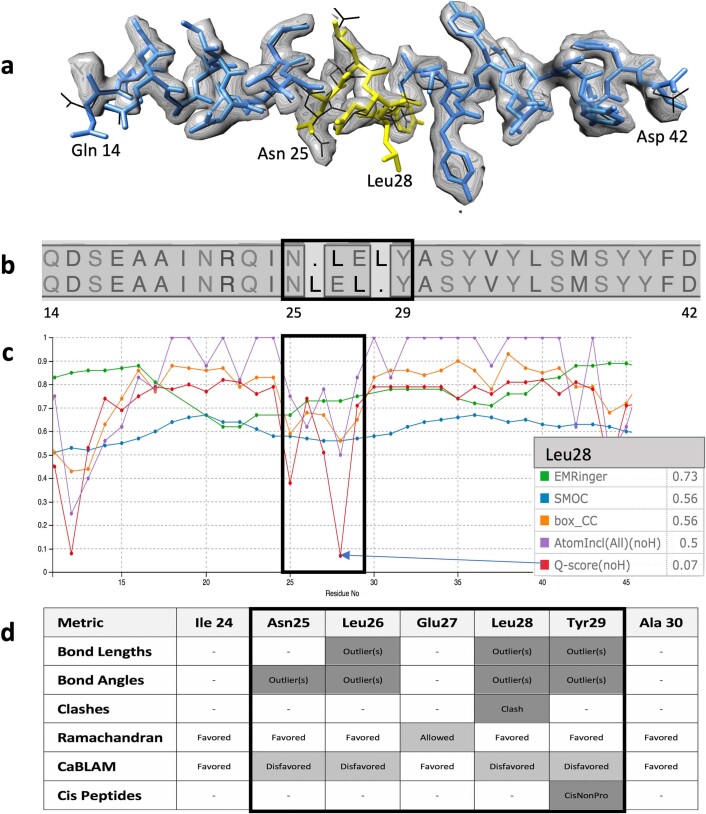
Extended Data Fig. 3. Evaluation of a short sequence misalignment within a helix.
Local Fit-to-Map and Coordinates-only scores are compared for a 3-residue sequence misalignment inside an ɑ-helix in an ab initio model submitted to the Challenge (APOF 2.3 Å 54_1). a, Model residues 14–42 vs target map (blue: correctly placed residues, yellow: mis-threaded residues 25–29, black: APOF reference model, 3ajo). b, Structure-based sequence alignment of the ab initio model (top) vs. reference model (bottom). c, Local Fit-to-Map scores (screenshot from Challenge model evaluation website Fit-to-Map Local Accuracy tool). Curves are shown for Phenix Box_CC (orange), EMDB Atom Inclusion (purple), Q-score (red) EMRinger (green), and SMOC (blue). The score values for model residue Leu 28 are shown in the box at right. d, Residue scores were calculated using the Molprobity server. The mis-threaded region is boxed in (b-d). Panels (a) and (b) were generated using UCSF Chimera.