Figure 1. Structure and topology of NMDARs.
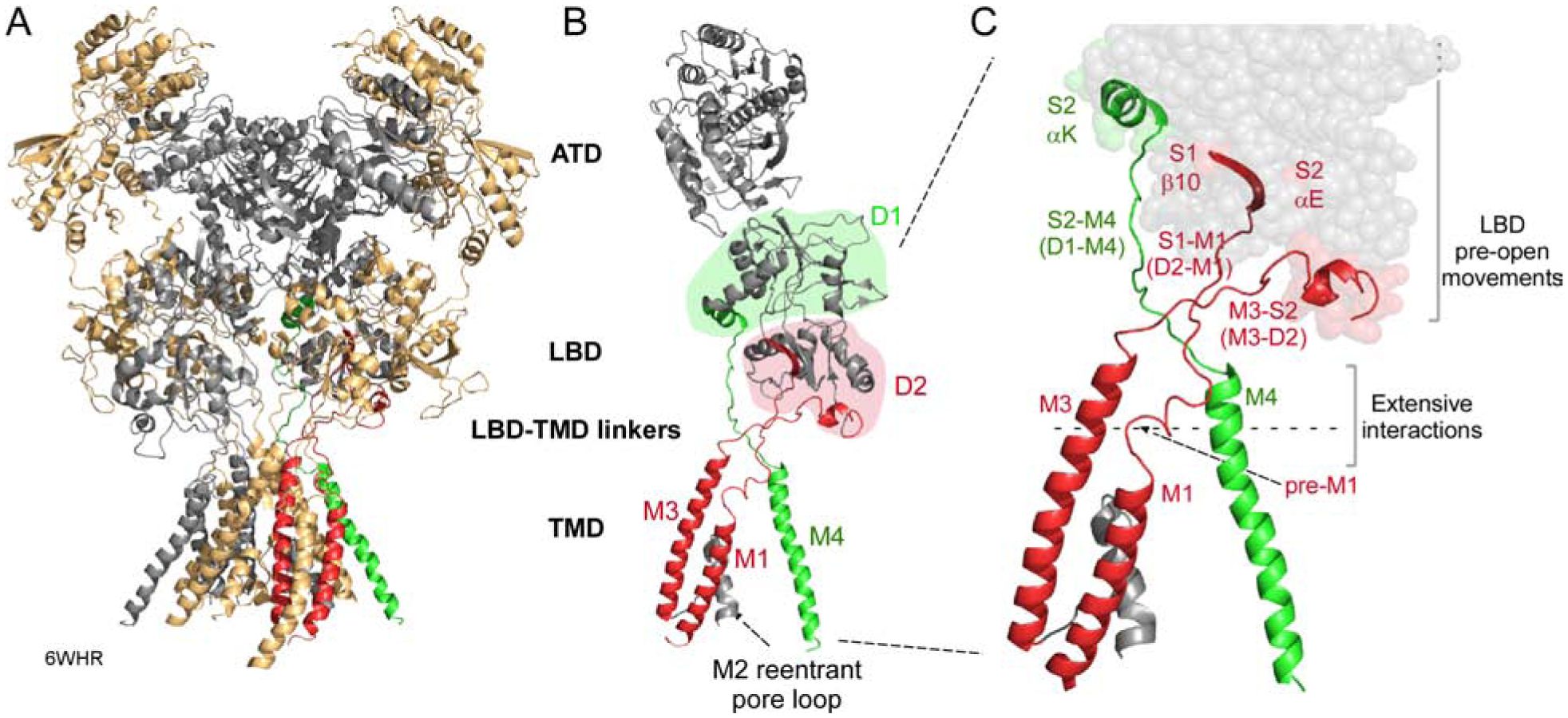
(A) NMDARs consist of four modular domains: extracellular amino-terminal (ATD) and ligand-binding (LBD) domains; the transmembrane domain (TMD) forming the ion channel; and an intracellular C-terminal domain (CTD). Subunits are colored light orange (GluN1) and gray (GluN2B). 6WHR (Chou et al., 2020).
(B) Topology of an individual subunit. LBD lobes are colored green (D1) or red (D2). The ion channel is formed by 3 transmembrane segments, M1, M3, and M4, and an intracellular M2 pore loop. Transmembrane segments are colored according to the LBD lobe to which they are connected.
(C) Bridging the LBD and TMD in each subunit are three LBD-TMD linkers: S1-M1 (D2-M1), M3-S2 (D2-M3), and S2-M4 (D1-M4). The most membrane proximal secondary structures in the LBD are: β10 (S1-M1); αE (M3-S2); and αK (S2-M4). With agonist binding these structures undergo movements that precede channel opening (Zhu et al., 2016, Tajima et al., 2016, Twomey et al., 2017, Chen et al., 2017a, Chou et al., 2020). Pre-M1 is a short helix in S1-M1 that surrounds the corresponding M3 segment (Sobolevsky et al., 2009).
