Figure 3. Nanoproteomics enables proteoform-resolved analysis of low-abundance cardiac troponin I in human serum.
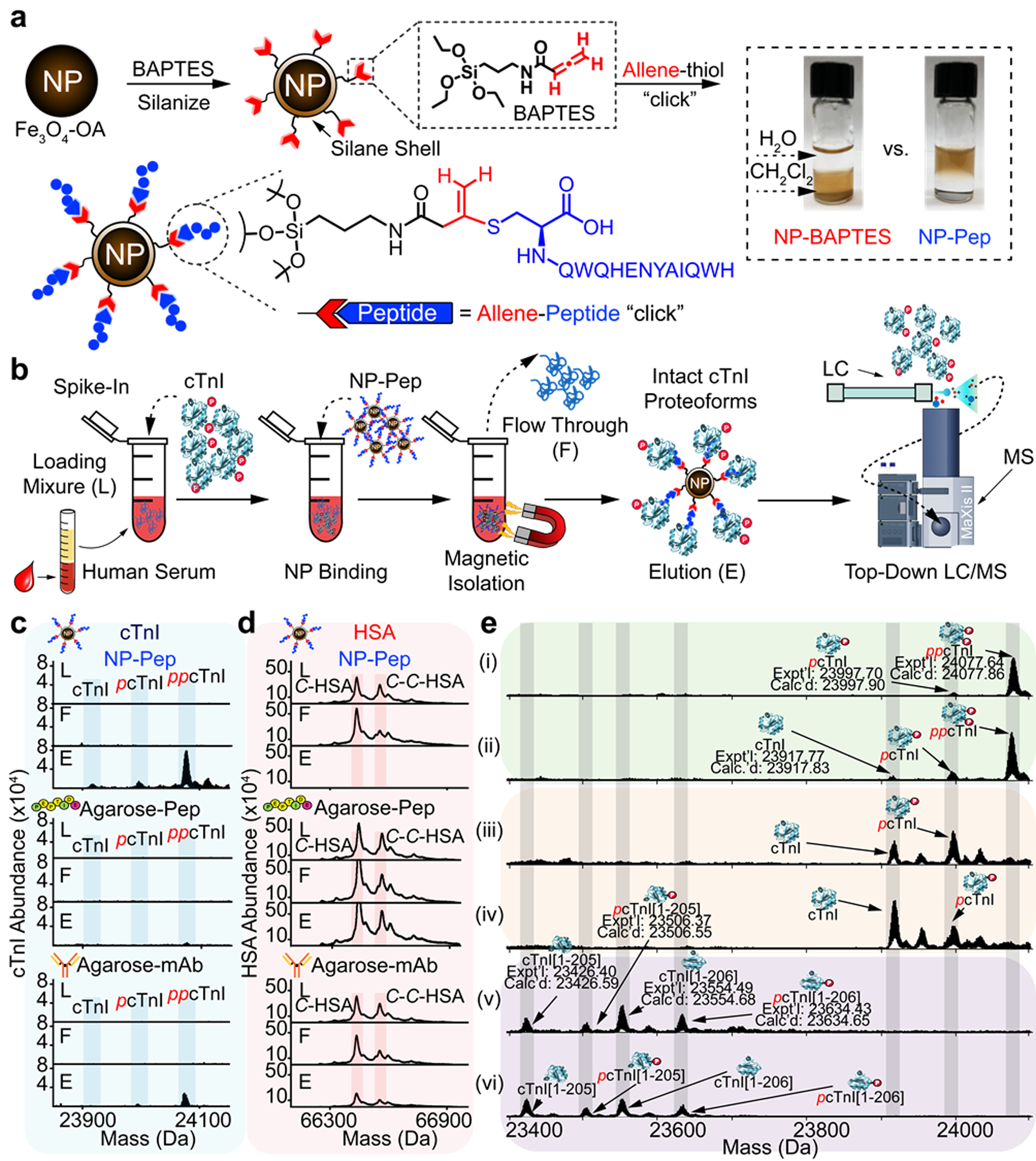
a, Silanization of Fe3O4 NPs using an allene carboxamide-based organosilane monomer (BAPTES) for cysteine thiol-specific bioconjugation. The rationally designed NPs are surface-functionalized with a 13-mer peptide that has a high affinity for cTnI (NP-Pep) for cTnI enrichment. The 13-mer peptide possesses a C-terminal cysteine that selectively reacts with the allene carboxamide moiety on the silanized NPs. The photograph shows functionalized NPs in a biphasic mixture of dichloromethane (CH2Cl2) and water (H2O), comparing the solvent compatibility of the NP-BAPTES and the NP-Pep. b, Nanoproteomics assay utilizing NP-Pep for specific enrichment of cTnI from serum and subsequent top-down MS analysis of cTnI proteoforms. cTnI is first spiked into human serum to prepare the loading mixture (L). The NPs are then incubated with the serum loading mixture, the cTnI-bound NPs are magnetically isolated, the unwanted and nonspecific proteins are removed as flow-through (F). The captured cTnI is then eluted and the final elution fraction after enrichment is analyzed by top-down LC/MS. c-d, Normalized deconvoluted mass spectra corresponding to enriched cTnI (c) and depleted HSA (d), illustrating the abundance of cTnI and HSA before and after enrichment using NP-Pep, high-affinity peptide-functionalized agarose (Agarose-Pep), and antibody (mAb M46) functionalized with agarose (Agarose-mAb). e, Deconvoluted MS corresponding to cTnI proteoforms enriched from human serum. The cTnI (~10–20 ng/mL) spiked in the human serum (10 mg) were extracted from various human hearts: (i) and (ii), donor hearts; (iii) and (iv), diseased hearts with dilated cardiomyopathy, (v) and (vi), post-mortem hearts. cTnI proteoforms were identified using accurate intact mass measurement, using the most abundant mass based on the amino acid sequence of entry name TNNI3_human from the UniProtKB sequence database. p, phosphorylation. pp, bisphosphorylation. Figure adapted from reference [68]. Copyright 2020 Springer Nature.
