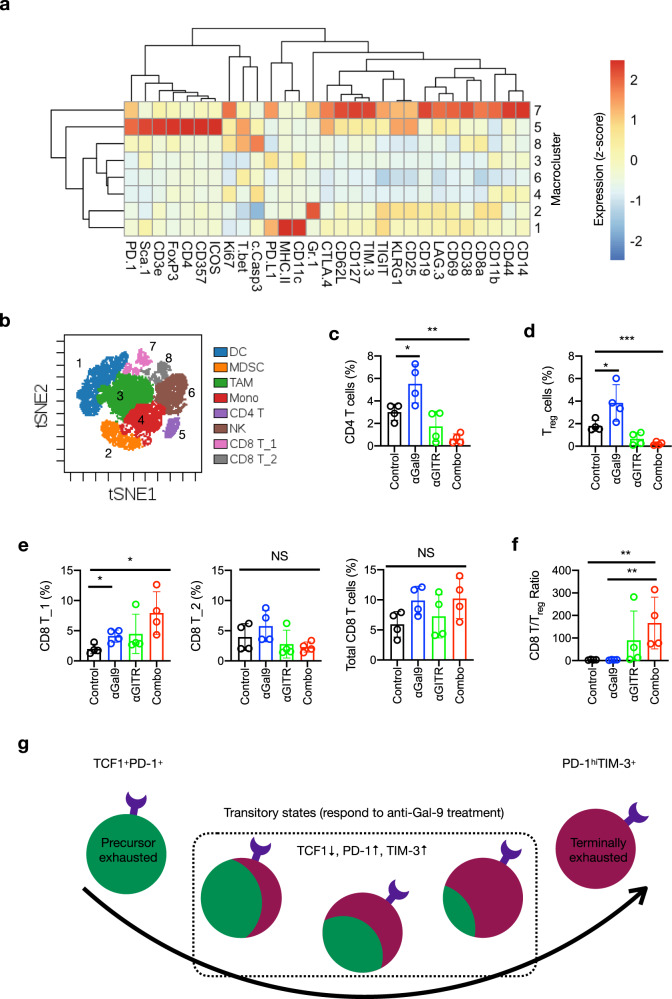
Fig. 6. Anti-Gal-9 therapy targets specific tumor-infiltrating T cell populations.
a Heatmap showing differential marker expression in CD45+ TIL clusters identified by analysis of CyTOF data using viSNE and FlowSOM. b Annotation of TIL populations based on differential marker expression as shown in (a) and Supplementary Fig. 6. DC, dendritic cell; MDSC, myeloid-derived suppressor cell; TAM, tumor-associated macrophage; Mono, monocytes; CD4 T, CD4 T cell; NK, natural killer cell; CD8 T_1, CD8 T cell subset 1; CD8 T_2, CD8 T cell subset 2. c–e T cell subset frequency in total CD45+ TILs. c CD4 T cells. Control vs αGal-9, P = 0.0337; control vs combo, P = 0.0013. d Treg cells. Control vs αGal-9, P = 0.0327; control vs combo, P = 0.0009. e CD8 T cell subsets. CD8 T_1 subset: control vs αGal-9, P = 0.0155; control vs combo, P = 0.0165. f CD8 T/Treg ratios in TILs from indicated treatment groups. Control vs combo, P = 0.0022, αGal-9 vs combo, P = 0.0021. g Proposed model of Gal-9 inhibition-elicited T cell response in tumor. The proliferating transitory T cells in the process of precursor exhausted T cells differentiation into terminally exhausted T cells are the major responders to anti-Gal-9 treatment. Unpaired two-tailed t tests were used for comparing means between treatment groups in (c–f). n = 4 mice in each treatment group. Error bars represent SD. Source data are provided as a Source data file.