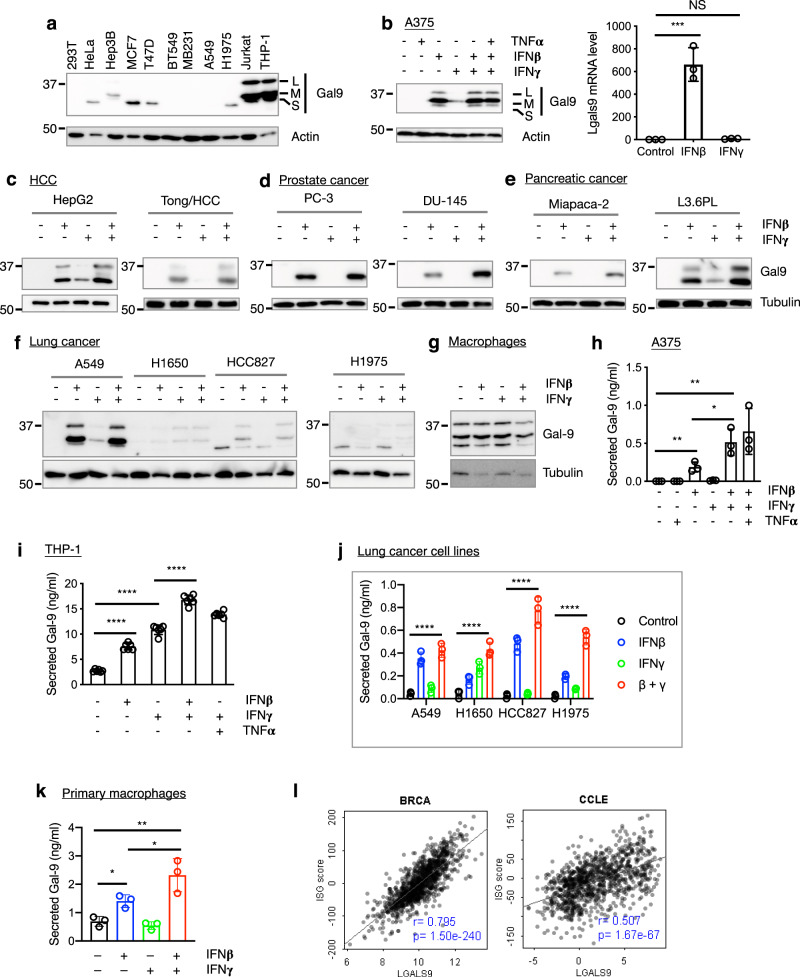
Fig. 7. Interferon β and γ promote Gal-9 expression and secretion.
a Baseline expression of Gal-9 protein in cancer cell lines. L, M, and S denote the three common Gal-9 isoforms resulting from alternative pre-mRNA splicing. b Regulation of Gal-9 expression in A375 human melanoma cells by indicated inflammatory cytokines at protein (left) or mRNA (right) levels. Data are presented as mean values ± SD. Statistical differences were assessed using ordinary one-way ANOVA with Dunnett’s multiple comparisons test. NS, not significant; ***P < 0.001. Control vs IFNβ, P = 0.0002; control vs IFNγ, P = 0.9940. c–g The effects of IFNβ and IFNγ on Gal-9 expression in cell lines of multiple cancer types and primary macrophages. h–k The effects of IFNβ and IFNγ on Gal-9 secretion from tumor cells and immune cells. h A375 melanoma cells. n = 3 independent experiments. Control vs IFNβ, P = 0.0074; IFNβ vs IFNβ + IFNγ, P = 0.0388; control vs IFNβ + IFNγ, P = 0.0070. i THP-1 monocytic leukemia cells. n = 6 independent experiments. Control vs IFNβ, P < 0.0001; control vs IFNγ, P < 0.0001; IFNβ vs IFNβ + IFNγ, P < 0.0001; IFNγ vs IFNβ + IFNγ, P < 0.0001. j Lung cancer cell lines. n = 3 independent experiments. Control vs IFNβ + IFNγ, P < 0.0001 for all the cell lines. k Primary macrophages. n = 3 independent experiments. Control vs IFNβ, P = 0.0117; IFNβ vs IFNβ + IFNγ, P = 0.0395; control vs IFNβ + IFNγ, P = 0.0099. l Expression correlation of LGALS9 with ISGs in the TCGA BRCA dataset and all cancer cell lines in CCLE, as analyzed by linear regression (Pearson correlation with two-tailed p-values). Unpaired two-tailed t tests were used for comparing means between treatment groups in (h–k). Each circle represents one experiment. Error bars represent SD. Data shown in (a–g) are representative of three independent experiments.