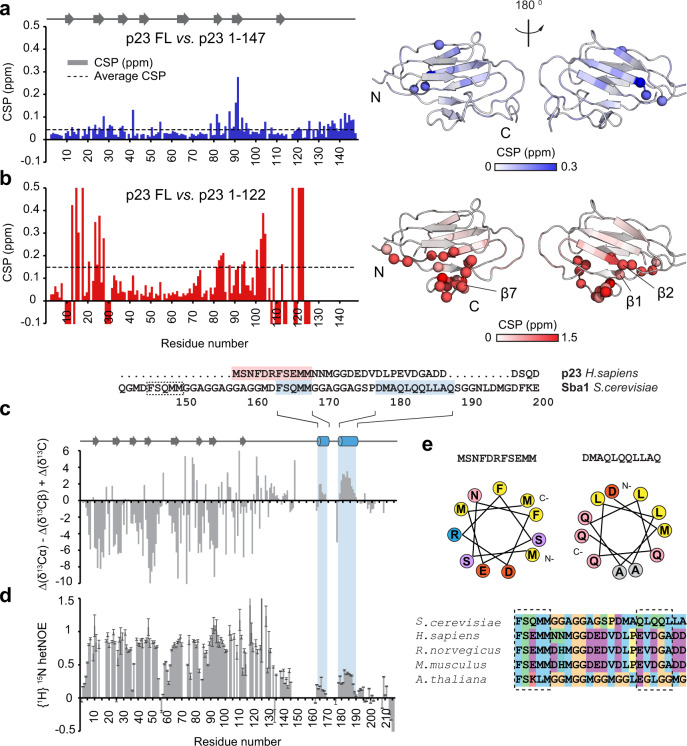
Fig. 3. NMR analysis of different constructs of p23.
Comparing chemical shift differences of the deletion mutants show that the p23Δ69 (1-147) mutant (a) is largely unaffected by the truncation of the tail. However, (b) the p23Δ94 mutant (1-122) reveals significant changes of the chemical shifts. Residues with a negative value indicate NMR signals which are absent in the NMR spectra of the tail-deleted construct. The spectral changes associated with tail deletions are plotted on the crystal structure of p23/Sba1 shown on the right. Color saturation indicates the strength of the chemical shift perturbation. Residues undergoing large chemical shift changes are shown as spheres. (c) Secondary 13C chemical shifts comparing differences in the Cα, Cβ, and C′ values versus random coil values of p23WT (1–216). Positive and negative secondary chemical shifts are indicative of α-helical and β-strand conformation, respectively. Secondary structure elements are indicated at the top and highlighted in blue for yeast. In addition, the sequence alignment of human p23 is included, with the helical motifs highlighted in blue (yeast) and red (human). An additional predicted helical motif in yeast is indicated by a dotted box. d {1H}-15N heteronuclear NOE values obtained from the ratio of peak intensities of saturated vs. non-saturated experiments. Values around 0.78 correspond to rigid regions. Lower values correspond to increasing flexibility at sub-nanosecond timescales. Errors were estimated from spectral baseplane noise RMSD according to Farrow et al.89. e Polarity distribution of identified helical segments reveals their amphipathic properties. Orange/yellow colors indicate hydrophobic residues, while red/pink/blue colors represent charged amino acids. The sequence alignment shown at the bottom indicates the conservation of helical motifs in the p23 tail across different species, colored according to the Clustal code.