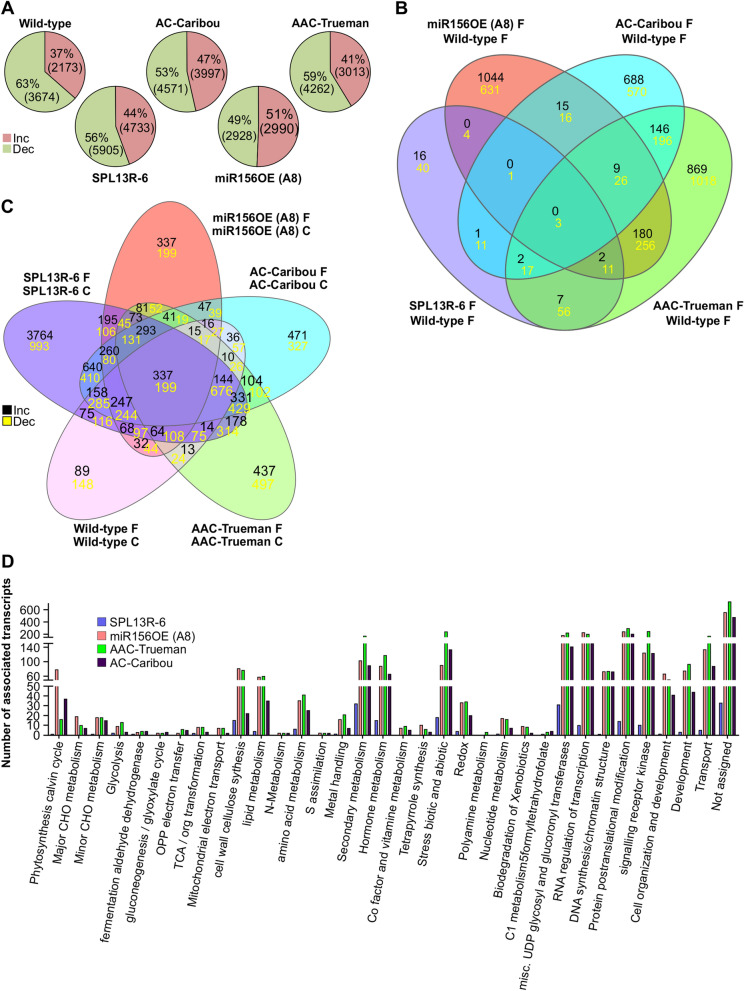
Figure 3.
Differentially expressed genes and their associated function upon flood stress in alfalfa. (A) Differentially expressed genes, DEG, in each genotype comparing flooding and well-drained control alfalfa plants, (B) Venn diagram illustrating commonality and uniqueness in DEG obtained by comparing flood-stressed WT to AC-Caribou, AAC-Trueman, SPL13RNAi-6 and miR156-A8 genotypes independently upon flooding, (C) Venn diagram illustrating commonly shared and unique DEG obtained by comparing flood-stressed and their respective well-drained control counterparts plants of each genotypes, (D) functional distribution of DEG generated by comparing flood-stressed WT to AC-Caribou, AAC-Trueman, SPL13RNAi-6 and miR156-A8 genotypes. Upper and lower panel numbers in ‘B’ and ‘C’ indicate increased and decreased DEG, respectively, compared to WT flood stress or the same genotype under control conditions respectively. Novaseq 6000-based RNAseq analysis was performed for three biological replicates for each treatment condition and genotype. Venn diagram was constructed using an online tool http://www.interactivenn.net/ from DEG.