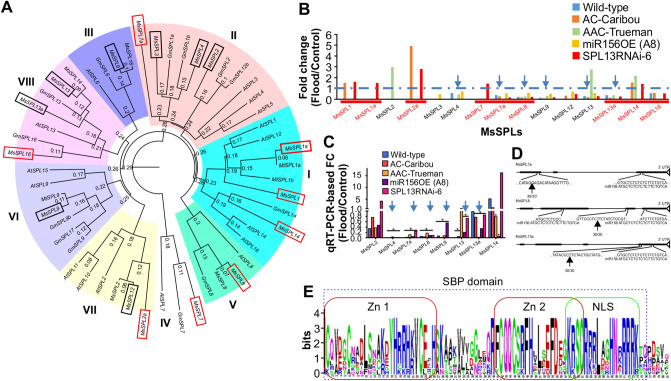
Figure 4.
Phylogenetic analysis and identification of new SPLs in alfalfa. (A) Amino acid coding sequence-based phylogenetic analysis of SPLs, (B) Differentially expressed SPLs between control and flood-stressed alfalfa plants, (C) Validation of the consistently regulated SPLs from RNAseq using qRT-PCR, (D) The conserved SQUAMOSA PROMOTER-BINDING PROTEIN, SBP, domain in the newly identified SPLs containing two zinc-finger binding domains (Zn 1 and 2) and nuclear localization signal (NLS), (E) Complementarity of SPL7a, 8 and 13a to that of matured miR156 sequence along with 5′ RLM RACE determined cleavage sites. SPLs boxed with red line in ‘A’ and underlined in ‘B’ are newly identified SPLs while boxed with black lines were previously identified (Aung et al., 2015b; Gao et al., 2016). Arrows in ‘B’ and ‘C’ indicates commonly reduced SPLs during FS. Phylogenetic analysis is done using clustal omega online tool (https://www.ebi.ac.uk/Tools/msa/clustalo/) followed by FigTree v.1.4.2 free software. The ‘*’ in ‘C’ indicates significance level at p < 0.05 when compared between control and flooding transcript levels. In silico amino acid sequence in ‘D’ is analysed using http://weblogo.berkeley.edu/logo.cgi. In ‘D’ zinc-finger binding domains (Zn 1 and 2) and nuclear localization signals (NLS) are indicated with red and green boxes, respectively. Arrows in ‘D’ indicates cleavage site by miR156.