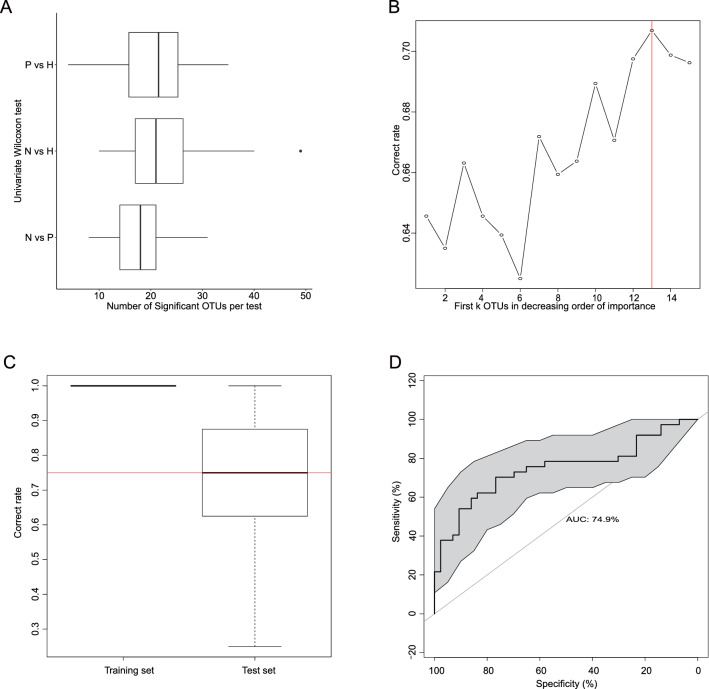
Figure 2.
Univariate comparison and machine learning based on HBeAg seroconversion. (A) Wilcoxon test is conducted to identify the significant genera in paired groups (P vs. H, N vs. H, N vs. P). This procedure is repeated 100 times on bootstrapped sample set. About 18–22 genera are selected. The number of significant genera for each comparison is reported with boxplot. (B) Thirteen genera are selected as microbial signature for HBeAg seroconversion to guarantee about 71% accuracy rate on test data. (C) The mean of accuracy rate on test data is about 0.75, indicating this method is robust. (D) The area under the ROC curve (AUC) is 74.9%, which indicates the prediction model has a significant discriminative ability.