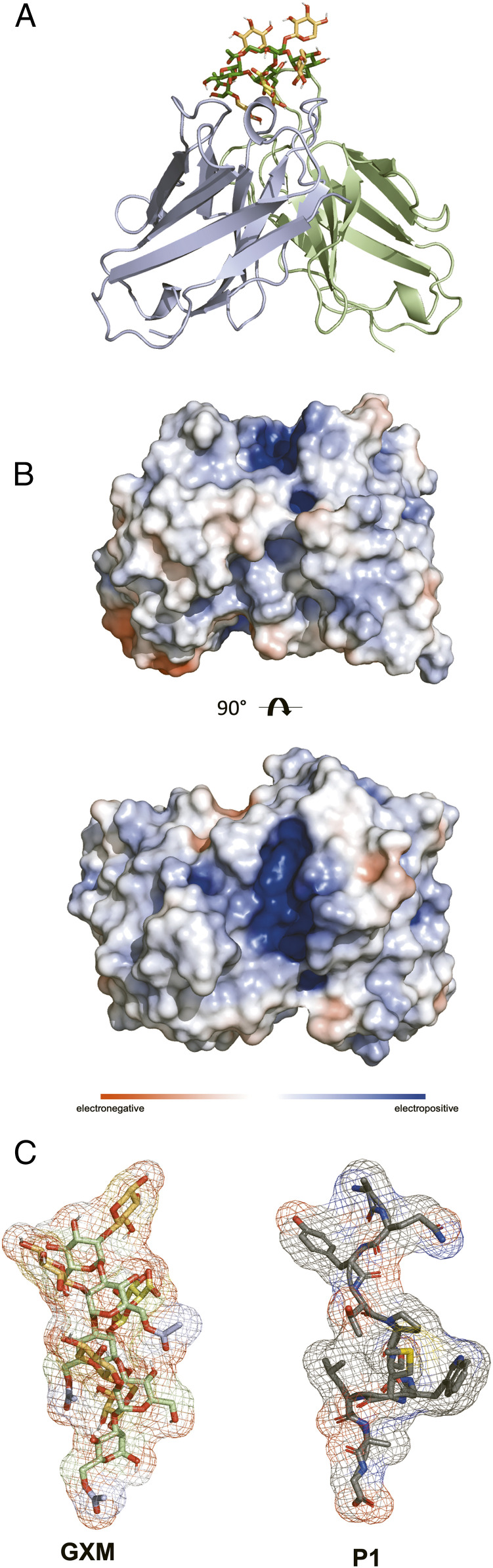
Fig. 4.
Crystal structure of 2H1 and GXM peptide mimotope P1 and molecular docking studies with GXM. (A) Crystal structure of anti-GXM mAb 2H1. VH shown in light blue and VL light chain shown in pale green with docked GXM. (B) Calculated electrostatic surface potential of 2H1. Electrostatic surface potentials are colored red for negative charges, blue for positive charges, and white color represents neutral residues. Electrostatic surface potential was calculated using adaptive Poisson–Boltzmann solve in PyMOL. mAb 2H1 shows a positively charged cleft (+10 kT to −10 kT). (C) GXM and P1 both adopt a helical conformation. Glycam Carbohydrate Builder was used to construct the ligand.