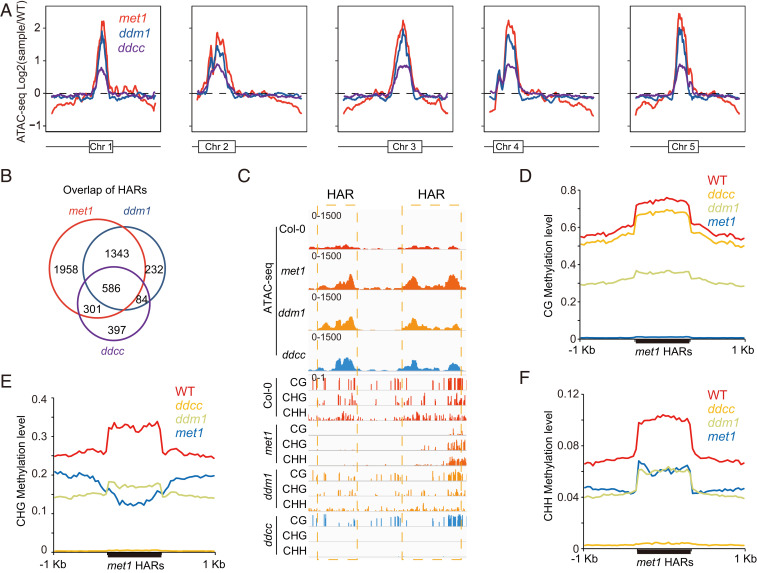
Fig. 3.
Connection between DNA methylation and chromatin accessibility. (A) Genome-wide pattern of chromatin accessibility variation in met1, ddm1, and ddcc. Variation in ATAC-seq signal (log2 mutant vs. Col-0) is depicted on the y axis. The box in each chromosome represents the pericentromeric heterochromatin region. (B) Overlap of peaks with more accessibility identified by ATAC-seq in met1, ddm1, and ddcc. (C) Screenshot showing simultaneous reduction of CG, CHG, and CHH methylations in peaks with more accessibility in met1, ddm1, and ddcc. Average distribution of CG (D), CHG (E), and CHH (F) methylation over peaks with more accessibility in met1.