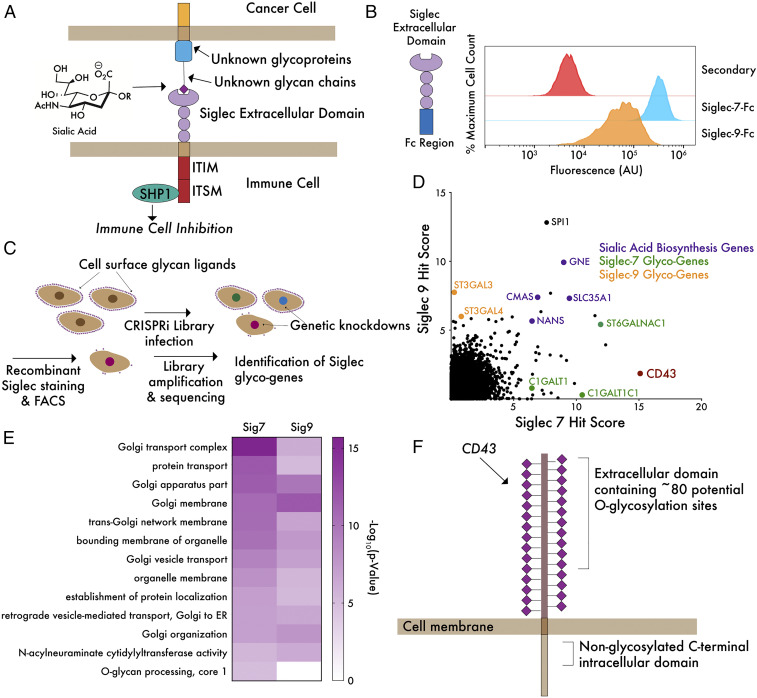
Fig. 1.
A genome-wide CRISPRi screen reveals core genes driving ligand expression for Siglec-7 and Siglec-9. (A) Siglecs possess an extracellular domain that binds to glycan ligands containing sialic acid. Ligand–receptor engagement initiates signaling through intracellular ITIM/ITSM domains, inhibiting immune activation. Sialic acid is incorporated into hundreds of glycan structures on thousands of individual proteins and lipids, making it challenging to define molecular details of ligand–receptor interactions. (B) K562 cells were incubated with 1 μg/mL Siglec-7–Fc and Siglec-9–Fc precomplexed to an Alexa Fluor 488 anti-huIgG secondary antibody and subjected to live cell flow cytometry. “Secondary” indicates noncomplexed Alexa Fluor 488 anti-huIgG. (C) A genome-wide library of ∼104,000 sgRNAs (five sgRNAs/gene) was transduced into K562-dCas9–KRAB cells and incubated with Siglec-7–Fc or Siglec-9–Fc as in B. FACS was performed to isolate a population of cells exhibiting at least a 10-fold reduction in ligand staining. Library amplification and sequencing was then performed to identify sgRNAs enriched in the low-staining population. (D) Hits for Siglec-7 and Siglec-9 were plotted and ranked by hit score (−log10[positive selection score]), where a higher value indicates a stronger enrichment of sgRNAs in the low-staining population. The screen identified a number of sialic acid biosynthesis genes (purple), glycotransferase genes specific for Siglec-7 and Siglec-9 (green and orange), as well as a single-cell surface glycoprotein, CD43, specific for Siglec-7 (red). (E) Enriched GO terms for hits in both screens show importance of Golgi and sialic acid biosynthesis genes. O-glycan biosynthesis is an enriched term in the Siglec-7 but not Siglec-9 screen. (F) The top hit of the Siglec-7 screen, CD43, is a mucin-type O-glycoprotein bearing many sialoglycan modification sites.