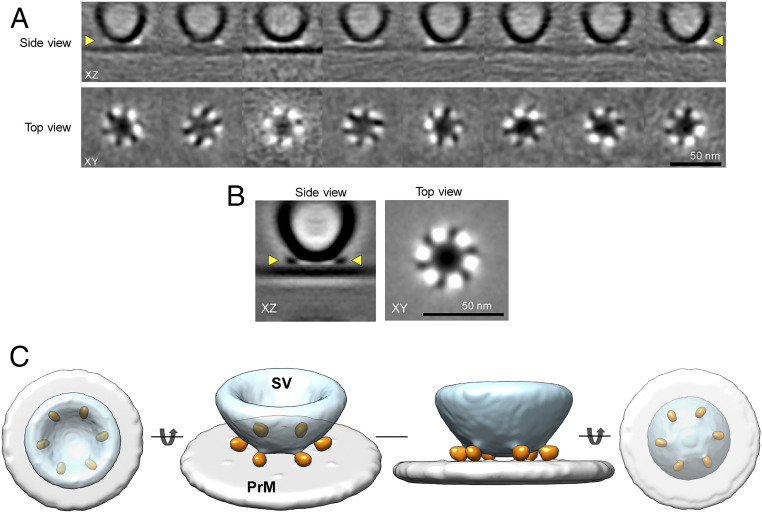
Fig. 4.
Symmetrical arrangement of protein density under docked synaptic vesicles. (A) Subtomogram class averages of SVs that are docked to the PrM. All the eight 3D class averages revealed a symmetrical organization of proteins at the SV–PrM interface. Side-view vesicles, with SV–PrM interbilayer distance of less than ∼6 nm and top/bottom-view vesicles, with a diameter of 43.88 ± 5.32 nm were pooled together for the subtomogram analysis. Top row: Slice through the center of the tomogram along the z axis. Bottom row: Corresponding slices through the volume in the XY plane at the vertical position highlighted by yellow arrowheads on the Top. (B) The cryo-ET 3D reconstruction of the SV–PrM interface reveals a symmetrical organization of six protein densities at the SV–PrM interface. (Left) Slice through the reconstructed volume along the z axis of the subtomogram averaged structure (XZ slice, side view) revealed protein densities connecting the SV to the PrM. (Right) Slice through the volume in XY plane (top view) at the vertical position highlighted by yellow arrowheads in the XZ slice revealed the symmetrical arrangement of protein densities at the SV–PrM interface. (C) Surface representation of the cryo-ET 3D reconstruction of the protein organization at the SV–PrM interface at different orientations. The cryo-ET 3D map at threshold level σ = 0.85 was segmented using UCSF Chimera for the surface representation. The top, side, and bottom views of the 3D reconstructions reveal six symmetrically organized protein densities (orange) connecting the SV (light blue) to the PrM (light gray).