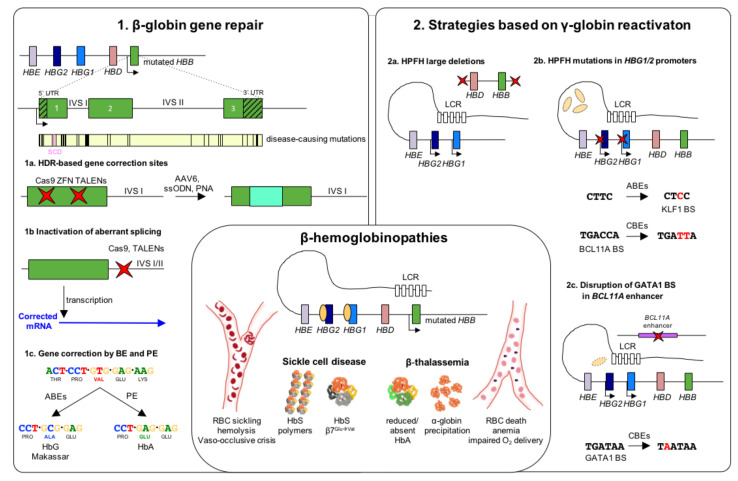
Figure 1.
Genome editing strategies for β-hemoglobinopathies. β-hemoglobinopathy-associated variants in the β-globin locus cause the insufficient (β-thalassemia) or abnormal (SCD) expression of β-globin. Reduced β-globin expression leads to globin imbalance and α-globin precipitation causing RBC death, anemia and impaired oxygen delivery. In SCD, a single amino acid substitution (β7Glu→Val) confers to the hemoglobin (HbS) a higher propensity to polymerize under hypoxic conditions, leading to RBC sickling, hemolysis and vaso-occlusive crises. (1). Strategies based on β-globin gene repair. Schematic representation of the β-globin gene including 5′ and 3′ untranslated region (UTR) and a map of the most common β-hemoglobinopathy-causing mutations. (1a) Several genome editing approaches have been tested for repairing β-globin mutations through the homology direct repair (HDR) pathway. (1b) Non-homologous end-joining (NHEJ) has been used to target aberrant splicing sites in introns I and II, resulting in the restoration of functional β-globin transcripts. (1c) Newly-established base (BE) and prime editing (PE) approaches proved their efficacy in correct the SCD-causing mutation. In particular, adenine base editors (ABEs) are suitable to convert the GTG codon into a GCG triplet, leading to the production of a non-pathogenic Hb variant (HbG Makassar). Thanks to a pegRNA-guided reverse transcriptase activity, PE can revert the SCD mutation, restoring the production of WT hemoglobin (HbA). (2). Strategies based on γ-globin reactivation. (2a) Nuclease-mediated strategies aimed at reproducing natural occurring large deletions that reactivate γ-globin expression. These deletions could either eliminate HbF inhibitory sequences or juxtapose the γ-globin promoters to remote enhancer regions. (2b) Targeting the HBG1/2 promoters with nucleases results in the disruption of binding sites (BS) for γ-globin transcriptional repressors, leading to a potent HbF expression. ABEs and cytosine base editors (CBEs) can be used to reproduce HPFH mutations that either create BSs for transcriptional activators (e.g., KLF1) or disrupt repressor BSs (e.g., BCL11A). (2c) Disruption of the GATA1 motif in the erythroid-specific intronic enhancer of BCL11A (achieved either using nucleases or CBEs) determines BCL11A knock-down and, consequently, reactivation of γ-globin expression. Red crosses indicate edits. LCR: locus control region; RBC: red blood cells; HbS, sickle hemoglobin; HbA, adult hemoglobin; IVS: intervening sequence; ZFN: zinc finger nuclease; TALENs: transcription activator-like effector nucleases; AAV6: adeno-associated viral vector serotype 6; ssODN: single-stranded donor oligonucleotides; PNA: peptide nucleic acid; HPFH: hereditary persistence of fetal hemoglobin.