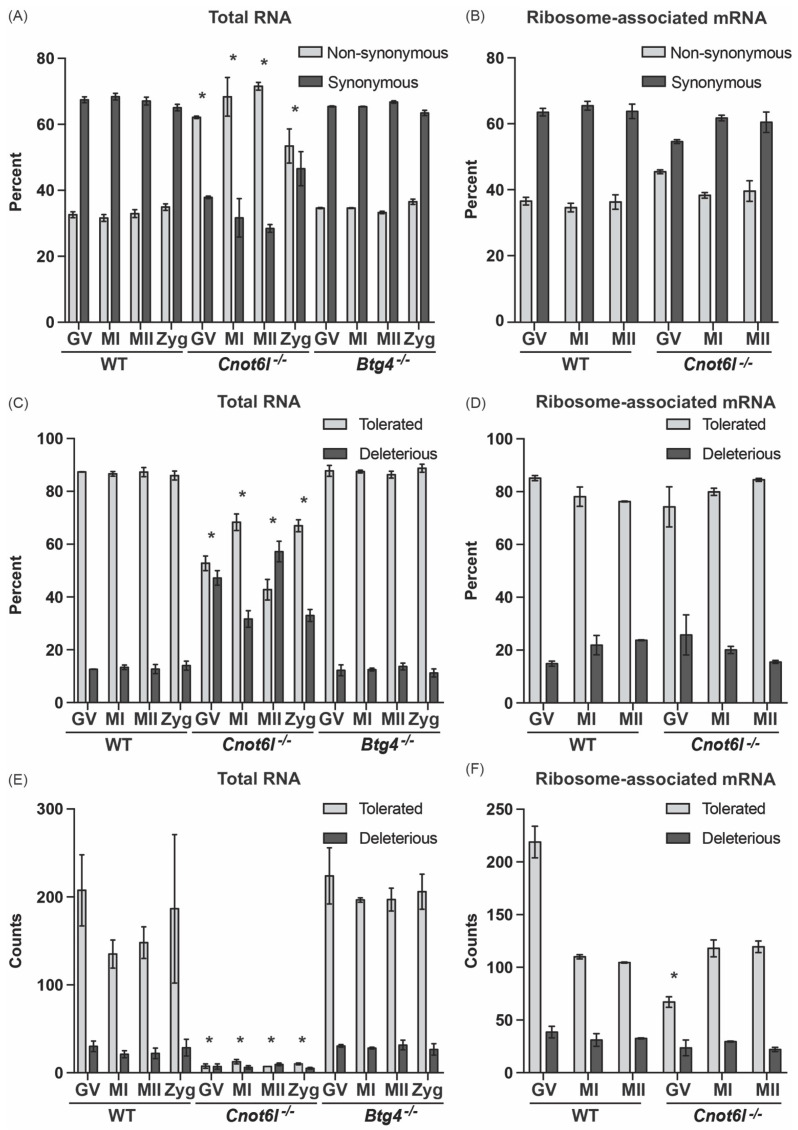
Figure 3.
Consequences of inosine RNA modifications within CDS from WT, Cnot6l-/-, and Btg4-/- oocytes, egg, and zygote transcripts. (A,B) Proportion of inosine modifications (percentage) in WT, Cnot6l-/-, and Btg4-/- samples that result in nonsynonymous or synonymous changes was determined for all CDS inosine modified mRNA transcripts in total RNA (A) and ribosome-associated mRNA (B). (C,D) Proportion of tolerated and deleterious transcripts following Sorting Intolerant From Tolerant (SIFT) analysis of the inosine modified mRNA transcripts from WT, Cnot6l-/-, and Btg4-/- samples in total RNA (C) or ribosome-associated mRNA (D). (E,F) Counts of tolerated and deleterious transcripts resulting from inosine RNA modifications from WT, Cnot6l-/-, and Btg4-/- samples in total RNA (E) or ribosome-associated mRNA (F). *Means ± SEM within a panel are different (p < 0.05); significance was determined using Χ2 tests or 2-way ANOVA. Only transcripts with TPM ≥ 1 were analyzed.