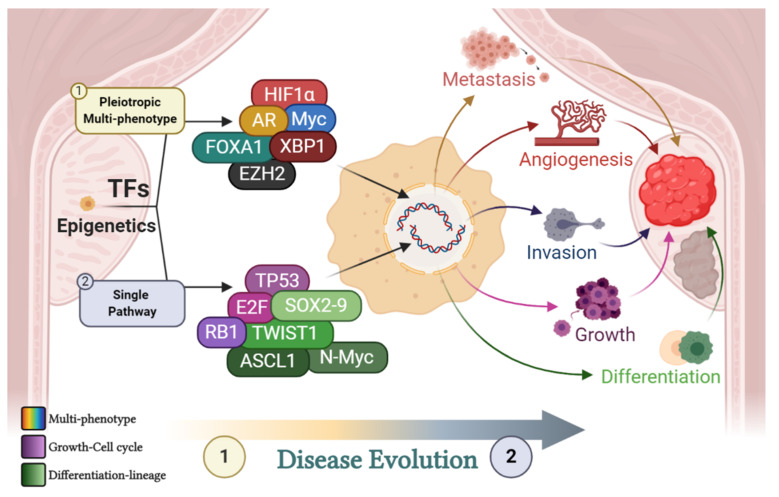
Figure 1.
Contribution of transcription factors (TFs) to disease progression. Multiple transcription factors and epigenetic modifiers contribute to disease progression. Some are pleiotropic and affect multiple key biological pathways (multi-colored). Others (although affecting several systems in a global way) predominantly affect single key developmental and biochemical pathways such as growth (purple array) and lineage determination or differentiation (green array). Moreover, pleiotropic TFs may be predominantly active at an early stage in disease evolution, whilst single pathway TFs are most prominent later in disease progression. This is exemplified by the phenomenon that localized prostate cancer (PCa) does not display a high proliferative index or huge fluctuations in the prostate-specific antigen (PSA) level prior to treatment (a marker of a transformed, differentiated prostate). As such, TFs are ever-present throughout disease evolution and collaborate to promote tumor differentiation, growth, angiogenesis, invasion, and metastasis. The purple array includes tumour protein 53 (TP53), E2 factor (E2F) and retinoblastoma protein (RB1). The green array includes the twist family BHLH transcription factor 1 (TWIST1), achaete-scute family BHLH transcription factor 1 (ASCL1), N-Myc and the sex determining region Y-box 2 (SOX) TF family. Individual TFs include the androgen receptor (AR), enhancer of zeste homologue 2 (EZH2), x-box binding protein 1 (XBP1), forkhead box A1 (FOXA1), Myc and hypoxia inducible factor 1 (HIF1alpha). Created with BioRender.com.