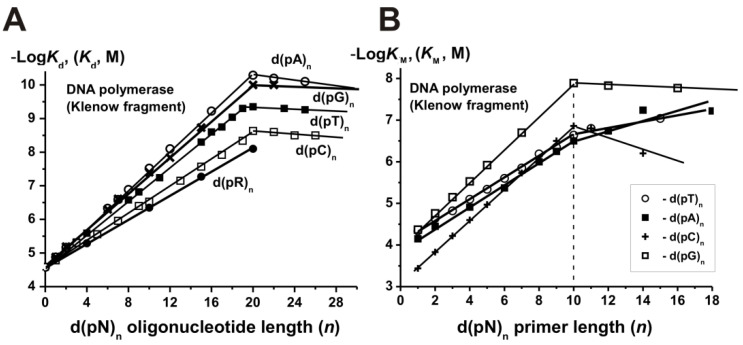
Figure 1.
Dependencies of −LogKd characterizing the affinity of ss d(pN)n deoxyribooligonucleotides to template-binding site of DNA polymerase (Klenow fragment, Escherichia coli) versus their length (n) (A). All d(pN)n are shown in panel A; d(pR)n oligomers do not contain any bases [19,20,21]. Dependencies of −LogKm values corresponding to different d(pN)n primers on their length (n) in the reaction catalyzing by Klenow fragment of E. coli. (B). The Km values in each case were determined using the poly(N) templates complementary to the primers used [23]. Figure 1A shows that the minimal ligands of the DNA polymerase template binding site are orthophosphate and various dNMPs. With the lengthening of the ss d(pN)n, the affinity gradually increases up to 20 nucleotide units, which are covered by the enzyme globule. In addition, the growth in the affinity of ss d(pN)n for the enzyme increases with an increase in the hydrophobicity of their bases: C < T < G < A. From the slopes of the curves for oligonucleotides, one can calculate the coefficient reflecting the increase in affinity with lengthening of each of the d(pN)n by one nucleotide unit.