Abstract
Recently, aptamers have attracted attention in the biosensing field as signal recognition elements because of their high binding affinity toward specific targets such as proteins, cells, small molecules, and even metal ions, antibodies for which are difficult to obtain. Aptamers are single oligonucleotides generated by in vitro selection mechanisms via the systematic evolution of ligand exponential enrichment (SELEX) process. In addition to their high binding affinity, aptamers can be easily functionalized and engineered, providing several signaling modes such as colorimetric, fluorometric, and electrochemical, in what are known as aptasensors. In this review, recent advances in aptasensors as powerful biosensor probes that could be used in different fields, including environmental monitoring, clinical diagnosis, and drug monitoring, are described. Advances in aptamer-based colorimetric, fluorometric, and electrochemical aptasensing with their advantages and disadvantages are summarized and critically discussed. Additionally, future prospects are pointed out to facilitate the development of aptasensor technology for different targets.
Keywords: aptamer, colorimetric aptasensor, fluorometric aptasensor, electrochemical aptasensor
1. Introduction
Owing to the advances in various areas of life, the population has become more exposed to pollutants, whether from industry, agriculture, or medical waste, resulting in an increase in the pollution associated with humans’ daily lives, leading to an increase in the rate of diseases. In this context, the development of fast, simple, low-cost, high-sensitivity, and specific sensors for detecting pollutants or early stages of diseases is important.
Because of the great advances in molecular biology and genetic engineering, the use of RNA and DNA has expanded not only in biology, for storing and transmitting genetic information, but also in the identification of antibiotics, proteins, peptides, amino acids, and even small molecules. Gold et al. and Szostak et al. reported the first studies involving a specific targeting affinity toward proteins [1,2].
Aptamers consist of 3D-folded structures of single-stranded oligonucleotides with lengths of usually 20–60 bases of nucleotides selected in vitro via the systematic evolution of ligand exponential enrichment (SELEX) process. The SELEX process is applicable for single targets, complex target structures, or even mixtures without proper knowledge of their composition. SELEX can be used to select aptamers with high affinities and specificities for their targets and low dissociation-constant values, across the low nanomolar to picomolar range. Aptamers can be selected through the in vitro process independent of animals or cell lines. The selection of aptamers for toxic target molecules or molecules with no or low immunogenicity is possible. Different modifications can be introduced in the basic SELEX process for the selection of the desired aptamer specifications for a specific application. The aptamers can be functional for native conformations of target molecules on live cells, so cell surface transmembrane proteins can be considered as targets [3,4,5,6].
The 3D folded structure permits the formation of stable complexes with various targets, such as proteins, nucleic acids, and small molecules [7,8,9,10,11,12,13]. Compared to antibodies, these aptamers are characterized by numerous advantages such as a wide range of targeted molecules (inorganics, organics, cells, viruses, and bacteria, among others), facile preparation on a massive scale, low molecular weights, high temperature and pH stability, easier modification, and long-term storage stability. These advantages promote their importance and application in many fields such as biosensing, therapy, and diagnostics [14,15,16,17,18,19,20,21,22,23].
Because of their size-dependent physical and chemical properties, biocompatibility, electronic properties, magnetic properties, and ability to manifest biological signaling and transduction mechanisms, the nanomaterials are providing great advances in the field of sensors. Several nanomaterials have been synthesized with different sizes and shapes such as gold nanoparticles (AuNPs), silica nanoparticles, silver nanoparticles (AgNPs), magnetic nanoparticles (MNPs), and carbon quantum dots (CQDs) as unique templates for many applications such as biomaterial assays; the diagnosis, monitoring, and treatment of disease; and drug delivery [24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. Interestingly, nanomaterials are being used as potential candidates for immobilization using aptamers to obtain nanosensor probes for several targets, with more amplification and various signals such as colorimetric, fluorometric, electrochemical, or optical [31,39,40,41,42,43,44,45,46,47,48,49]. Sensor fabrication mainly depends on target recognition and signal transduction. Thus, nanomaterials provide desirable signal transduction for converting molecular recognition events into physically detectable fluorescent, colorimetric, and electrochemical signals [50,51]. Simultaneously, nanomaterials play a significant role regarding an increase in the immobilization density and orientation of aptamers due to the high surface areas of nanomaterials and, thus, provide high binding capacity for targets [52,53,54]. However, the high accumulation leads to the restriction of the 3D-structure formation. Because of that, the aptamer density on the nanomaterial surface needs to be precisely optimized [55,56].
This systematic review is focused on recent approaches to aptamer engineering for biorecognition for several objectives. Herein, previous and current advances related to aptamer-based sensing protocols are provided, highlighting the possible detected signals, with a focus on the use of different nanomaterials with distinct configurations. The most significant studies on aptasensor development have been collected, providing the possible strategies available for using aptasensors. Moreover, a deep discussion on colorimetric, fluorescence, and electrochemical detection strategies is provided.
2. Colorimetric Aptasensor
Among the available assay recognition signals, colorimetric methods are considered simple and efficient with a great potential for point-of-care diagnostics, as the detection responses are simply visually discerned by the naked eye using simple, low-cost, and effective instrumental techniques. The basic colorimetric strategies involve the detection of the target by a color change through the naked eye and simple instrumentation. Colorimetric biosensors based on aptamers have demonstrated their sensitivity and selectivity, in addition to their effective potential for rapid onsite diagnosis without complicated instrumentation. However, the colorimetric aptasensor has some limitations such as the influence of color from samples; the time-consuming nature of the fabrication process [57,58]; the difficulty in employing it in multiple-target assays, which are highly demanded in clinical diagnosis [59]; and the small range of optimized pH solutions [60].
Nobel metal nanomaterials, in particular, AuNPs and AgNPs, are excellent signal transducers for colorimetric analysis due to their significant optical properties associated with their particle size, size distribution, and shape [61,62,63]. The intrinsic surface plasmon resonance (SPR) properties of AuNPs and AgNPs contribute significantly to colorimetric signal generation [64]. AuNPs are used for color-change-based detection, in which the AuNPs are employed as nanoassembly units for immobilization with aptamers for constructing a colorimetric aptasensor because of their unique features, including simple synthesis and unique optical, thermal conductivity, and electronic properties [65,66,67,68]. Moreover, AuNPs show excellent biocompatibility compared to AgNPs, which are known for their toxicity [69]. Furthermore, the large specific surface area facilitates numerous adsorptions of biomacromolecules onto AuNP surfaces via electrostatic interaction, protecting them against aggregation and rendering them a good signal transducer for aptasensor construction [70,71]. Based on the unique colorimetric capabilities of the distance-dependent surface plasmon resonance of AuNPs, a great variety of label-free colorimetric bioassay strategies have been developed. The color of AuNPs is extremely sensitive to their dispersion and aggregation in a solution, comprising the interparticle plasmon coupling changing, inducing SPR shifts [72]. However, there are some limitations in using a colorimetric sensor based on the SPR response other than the common disadvantages of colorimetric sensors previously mentioned, including the fact that the SPR sensors are prone to interference due to not only the absence of a response to a change in the refractive index, but also non-specific binding that can induce interference [73,74], leading to false results and limiting their applicability. In some cases, variations in the plasmonic response depending on the location of the target on the surface of the nanoparticles [75,76] are detected. Moreover, large nanoparticle aggregates are unstable in solution, which could lead to some measurement error [77,78,79].
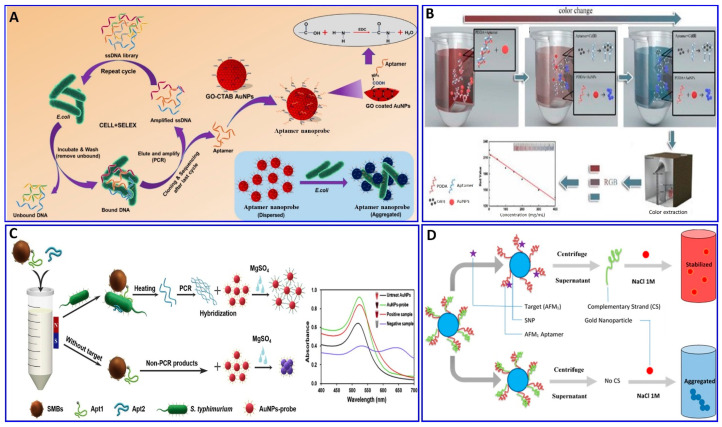
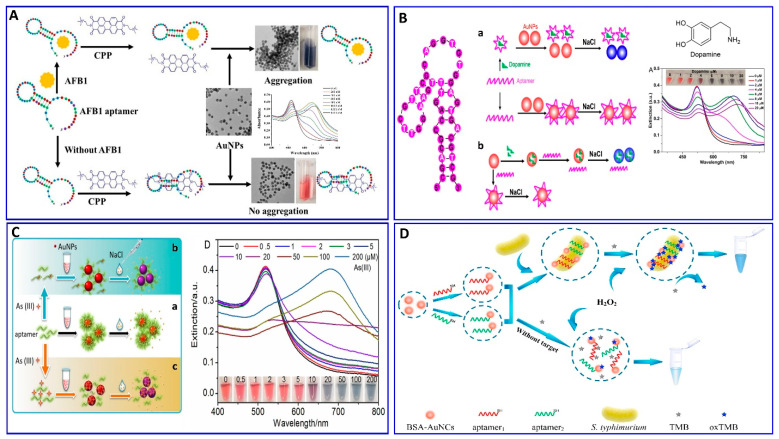
In this regard, the binding interaction induces a change in the refractive index around the surface of the AuNP that modulates its resonance angle. Therefore, the SPR has been used in an aptamer-based colorimetric strategy for the assay of several targets [80,81,82,83]. Gupta et al. proposed fast, easy, and cost-effective naked-eye visible detection for an Escherichia coli (E. coli) assay based on AuNPs’ aggregation as described in Figure 1A. In this assay, the AuNPs were covered with graphene oxide (GO), followed by functionalization with a specific aptamer for E. coli, establishing a stabilizing layer around the AuNPs. In the presence of the E. coli target, an aptamer–E. coli competitive interaction with AuNPs occurs, inducing the aggregation of AuNPs, with a distinct color change from red to blue and a low limit of detection (LOD) of 101 cells/mL, which is observed visually by the naked eye [84]. Moreover, Lai et al. assayed malathion via salt-induced AuNP aggregation with aptamer assistance for improving the sensor selectivity, where the designated aptamer for malathion was initially adsorbed on the surface of the AuNPs, enhancing their colloidal stability against aggregation in a highly saline solution. In the presence of malathion, the specific aptamer departs from the AuNP surface to bind with the malathion target, leaving the AuNP surface unprotected and tending to aggregate in a highly concentrated NaCl solution, with a distinct color change from red to blue. For more signal amplification, Lai et al. used the aggregated AuNPs as a promoter for catalyzing the Fehling reaction, producing different-size Cu2O with different resonance scattering intensities and a LOD of 5.24 ng/L (15.86 pM) [85]. AuNPs were functionalized with aptamers equipped with colorimetric smartphone technology for a sensitive and selective Cd2+ detection strategy based on the AuNP aggregation induced by polydiene dimethyl ammonium chloride (PDDA) [86], in which the selective aptamer for Cd2+ shows a stable complementary hybridization with the PDDA. In the presence of Cd2+, the aptamer leaves the PDDA and binds to the Cd2+; AuNP aggregation is induced by the aptamer leaving the PDDA, which induces the solution’s color change from red to blue, with a linear range of 1–400 ng/mL and a LOD of 1 ng/mL, using a smartphone device as indicated in Figure 1B. Chen et al. also used aptamer–AuN-induced aggregation under the effect of a highly saline solution for assaying viable Salmonella typhimurium (S. typhimurium) bacteria using a dual aptamer approach, in which one was functionalized with magnetic beads to capture S. typhimurium and the other was used for PCR amplification, as shown in Figure 1C [87]. Finally, the hybridization between the amplicons and the AuNP probe took place for 5 min before adding MgSO4 to induce the aggregation. The system showed a linear relationship for S. typhimurium from 3.3 × 101 to 3.3 × 106 CFU/mL, with LODs of 33 and 95 CFU/mL for detection by the naked eye in pure culture and milk, respectively (Table 1) [87]. S. typhimurium bacteria have also been detected via AuNP aggregation through competitive aptamer–target binding under the influence of salts, as inducers of aggregation, with a linear range of 100 to 109 CFU/mL and LOD of 16 CFU/mL, as reported by Jiecan [88]. The AuNP salt-induced aggregation mechanism was determined in an assay for aflatoxin M1 (AFM1) as described by Seyed et al. [89]. In this assay, a colorimetric aptasensor for the AFM1 assay was based on the protection of AuNPs against NaCl-induced aggregation via detaching the complementary strand of the aptamer (CS) from aptamer-modified streptavidin-coated silica nanoparticles (SNPs) in the presence of AFM1, as outlined in Figure 1D, with a linear dynamic range of 300–75,000 ng/L and LOD of 30 ng/L [89]. Similarly, a colorimetric aptasensor based on the aggregation of AuNPs as a signal transducer using cationic perylene probe as gold aggregating agent (CPP) was also provided by Lerdsrias et al. for assaying aflatoxin B1 (AFB1), with a LOD of 0.18 ng/mL [90], as shown in Figure 2A.
Figure 1.
(A) Schematic illustration of the colorimetric aptamer sensor for E. coli assay via target inducing aggregation of gold nanoparticles (AuNPs), reproduced from [84]. (B) Schematic illustration of the colorimetric aptasensor for Cd2+ assay based on AuNP aggregation, reproduced from [86]. (C) Schematic procedure for an S. typhimurium bacterium assay via a proposed colorimetric SMBs-Apt1 sandwich strategy, reproduced from [87]. (D) Schematic illustration of a colorimetric aptasensor assay for AFM1 based on salt aggregation of AuNPs induced by AuNP–aptamer competitive binding, reproduced from [89].
Table 1.
Examples of application of aptasensors for quantitative detection.
| Sensor Type | Target | Aptamer Sequence | Detection Range | LOD | Ref. |
|---|---|---|---|---|---|
| Colorimetric | E. coli | Fp: TAGGGAAGAGAAGGACATATGAT, Rp: TTGACTAGTACATGACCACTTGA) | 101–108 cells/mL | 101 cells/mL | [84] |
| Malathion | 5′-TAT ACA CAA TTG TTT TTC TCT TAA CTT CTT GAC TGC-3′ | 0–4000 ng/L | 5.24 ng/L | [85] | |
| Cd2+ | 5′-CTCAGGACGACGGGTTCACAGTCCGTTGTC-3′ | 1–400 ng/L | 1 ng/L | [86] | |
| Salmonella typhimurium | Apt1: 5′-biotin-GAGGAAAGTCTATAGCAGAGGAGATGTGTGAACCGAGTAA-3′ | 3.3 × 101–3.3 × 106 CFU/mL | 33 CFU/mL | [87] | |
| Apt2: 5′-CTCCTCTGACTGTAACCACGGAGTTAATCAATACAAGGCGGGAACATCCTTGGCGGTGCCGCATAGGTAGTCCAGAAGCC-3′ | |||||
| Salmonella typhimurium | NH2-GCGCTCGGCCTCCTCTGCCATCTCATTCGCGAGCGC | 100–109 CFU/mL | 16 CFU/mL | [88] | |
| AFM1 | 5-Biotin-ACTGCTAGAGATTTTCCACAT-3′ | 0.3–75 ng/mL | 0.03 ng/mL | [89] | |
| AFB1 | 5′-GTTGGGCACGTGTTGTCTCTCTGTGTCTCGTGCCCTTCGCTAGGCCCACA-3′ | 1–6 ng/mL | 0.18 ng/mL | [90] | |
| Salmonella typhimurium | Apt1 5′-AGT AAT GCC CGG TAG TTA TTC AAA GAT GAG TAG GAA AAG A-C6-SH-3′ | 101–106 CFU mL−1 | 1 CFU/mL | [97] | |
| Apt2 5′-TAT GGC GGC GTC ACC CGA CGG GGA CTT GAC ATT ATG ACA G-C6-SH-3′ | |||||
| ABA | AAAATGGGTTAGGTGGAGGTGGTTATTCCGGGAATTCGCCCTAAATACGAGCAAC | 1 nM to 10 μM | 0.51 nM | [98] | |
| Cortisol | 5′-GGA ATG GAT CCA CAT CCA TGG ATG GGC AAT GCG GGG TGG AGA ATG GTT GCC GCA CTT CGG CTT CAC TGC AGA CTT GAC GAA GCT T-3′ | 0.1–1000 nM | 0.1 nM | [99] | |
| Thrombin | TBA1 (5′-thiolated-TTT TTT TTT TTT TTT GGT TGG TGT GGT TGG-3′) | 0–10 μg/mL | 1.33 μg/mL | [100] | |
| TBA2 (5′-thiolated-TTT TTA GTC CGT GGT AGG GCA GGT TGG GGT GAC T-3′) | |||||
| Cd2+ | 5′-biotin-ACC GAC CGT GCT GGA CTC TGG ACT GTT GTG GTA TTA TTT TTG GTT GTG CAG TAT GAG CGA GCG TTG CG-3 | 1–500 ng/mL | 0.7 ng/mL | [101] | |
| PDGF-BB | 5′-CAGGCTACGGCACGTAGAGCATCACCATGATCCTG-3′ | 1–25 pM | 0.94 pM | [103] | |
| ATP | 5′-ACC TGG GGG AGT ATT GCG GAG GAA GGT-3′ | 0.50–100 μM | 0.09 μM | [106] | |
| Pb2+ | 5′-biotin-GGGTGGGTGGGTGGGT-3′ | 1–300 ng/mL | 0.63 ng/mL | [107] | |
| E. coli | Apt1: 5′-biotin-TGAGCCCAAGCCCTGGTATGCGGATAACGAGGTATTCACGACTGGTCGTCAGGTATGGTTGGCAGGTCTACTTTGGGATC-3′ | 16 to 1.6 × 106 CFU/mL | 2 CFU/mL | [108] | |
| Apt1: 5′-biotin-TGAGCCCAAGCCCTGGTATGAGCCCACGGAACACTGGTCGCGCCCACTGGTTTCTATATTGGCAGGTCTACTTTGGGATC-3′ | |||||
| 17β-E2 | 5′-GCTTCCAGCTTATTGAATTACACGCAGAGGGTAGCGGCTCTGCGCATTCAATTGCTGCGCGCTGAAGCGCGGAAGC-3′ | 1.5–50 nM | 1.5 nM | [109] | |
| AFB1 | 5′-biotin-GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCT TCG CTA GGC CCA CA-3′ | ND | 0.1 ng/mL | [110] | |
| PSA | 5′-Biotin-ATTAAAGCTCGCCATCAAATAGC-3′ | 0–2.1 ng/mL | 0.15 ng/mL | [111] | |
| E.coli O157: H7 | 5′-ATCCGTCACACCTGCTCTGTCTGCGAGCGGGGCG | 500–5 × 107 CFU/mL | 250 CFU/mL | [112] | |
| CGGGCCCGGCGGGGGATGCGTGGTGTTGGCTCCCGTAT-3′ | |||||
| Fluorometric | T-2 | 5′-CAGCTCAGAAGCTTGATCCTGTATATCAAGCATCGCGTGTTTACACATGCGAGAGGTGAAGACTCGAAGTCGTGCATCTG-3′ | 0.001−100 ng/mL | 0.57 pg/mL | [119] |
| DGX | 5′-AGCGAGGGCGGTGTCCAACAGCGGTTTTTTCACGAGGAGGTTGGCGGTGG-3′ | 0.001 to 0.5 ng/mL | 0.0032 ng/mL | [120] | |
| ZEN | 5′-NH2-AGCAGCACAGAGGTCAGATGTCATCTATCTATGGTACATTACTATCTGTAATGTGATATGCCTATGCGTGCTACCGTGAA-3′ | 31.4–628 nM | 7.5 nM | [121] | |
| PAT | 5′-GGC CCG CCA ACC CGC ATC ATC TAC ACT GAT ATT TTA CCT T-3′CFL | 5–300 ng/mL | 0.13 ng/mL | [130] | |
| AFB1 | TARMA-5′-GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCT TCG CTA GGC CCA CA-3′ | 0–180 ng/mL | 0.35 ng/mL | [123] | |
| AMP | 5′-CACGGCATGGTGGGCGTCGTG-Thiol-3′ | 100–1000 pM | 29.2 pM | [131] | |
| IFN-γ | Apt1: 5′-H2N-C6-CCGCCCAAATCCCTAAGAGAAGACTGTAATGAC ATCAAACCAGACACACACTACACACGCA-3′ | 2.0 × 10−18–5.0 × 10−8 M | 0.178 fM | [132] | |
| Apt2: 5′-TGGGGTTGGTTGTGTTGGGTGTTGTG-Azide(N3)-3′ | |||||
| MUC1 | 5′-Cy3-GCAGTTGATCCTTTGGATACCCTGG-NH2-3′ | 0–50 nM | 0.8 nM | [133] | |
| Isocarbophos | 5′-AGCT2GCTGCAGCGAT2CT2GATCGC2ACAGAGCT-3’ | 10–500 nM | 3.38 nM | [134] | |
| Hg2+ | 5′-FAM-TTC TTT CTT CCC CTT GTT TGT T-3′ | 20–150 nM | 15 nM | [135] | |
| E. coli | 5′-CCG GAC GCT TAT GCC TTG CCA TCT ACA GAG CAG GTG TGA CGG-C6 NH2-3′ | 85 to 85 × 107 CFU/mL | 17 CFU/mL | [140] | |
| E. coli ATCC8739 | 5′-ATCCGTCACACCTGCTCTGTCTGCGAGCGGGGCGCGGGCCCGGCGGGGGATGCGTGGTGTTGGCTCCCGTAT-3′ | 58–58 × 106 CFU/mL | 10 CFU/mL | [141] | |
| Malathion | 5′-ATCCGTCACACCTGCTCTTATACACAATTGTTTTTCTCTTAACT TCTTGACTGCTGGTGTTGGCTCCCGTAT-3′ | of 0.01–1 μM | 1.42 nM | [142] | |
| Pb2+ | Apt1: 5′-Biotin-CGA TCA CTA ACT ATr AGG AAG AGA TG-HS-3′ | 25–1400 nM | 5.7 nM | [143] | |
| Apt2: 5′-NH2-TGA GTG ATA AAG CTG GCC GAG CCT CTT CTC TAC-3′ | |||||
| Chlorpyrifos | 5′-CCTGCCACGCTCCGCAAGCTTAGGGTTACGCCTGCAGCGATTCTTGATCGCGCTGCTGGTAATCCTTCTTTAAGCTTGGCACCCGCATCGT-3′ | 5–600 nM | 3.8 nM | [144] | |
| ATP | 5′-CCCCAACTCCTTCCCGAAACCTACCTGGGGGAGTATTGCGGAGGAAGGTTTCGGG-3′ | 20–220 μM | 0.38 μM | [145] | |
| ATP | 5′-CCCCCCCCCCCCCACCTGGGGGAGTATTGCGGAGGAAGGT-3′ | 50 pM–1.0 nM | 26 pM | [152] | |
| AFB1 | 5′-GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCT TCG CTA GGC CCA CA-3′ | 2–400 ng/mL | 1.82 ng/mL | [153] | |
| Acetamiprid | 5′-TGT AAT TTG TCT GCA GCG GTT CTT GAT CGC TGA CAC CAT ATT ATG AAG A-3′ | 5 nM–1.2 μM | 3 nM | [154] | |
| Exosomes | 5′-CATCCATGGGAATTCGTCGACCCTGCAGGCATGCAAGCTTTCCCTATAGTGAGTCGTATTACTGCCTAGGCTCGAGCTCG-3′ | 0.68–30.4 pM | 0.57 pM | [121] | |
| ATP | 5′-FAM-AATTCTGGGGGAGCCTTTTGT GGG TAGGGC GGG TTG GTT TTG CCC CGG AGG AGG AATT-BHQ1-3′ | 1−500,000 μM | ND | [159] | |
| Cd2+ | 5′-AGTGACGTGCTGGACTCCGGACTATTGTGGTATGATCTGGTTGTGACTATGCAGTGCGTGCA-(CH2)3-SH-3′ | 20 pM–12 nM | 1.2 pM | [161] | |
| MC-LR | 5′-GGC GCC AAA CAG GAC CAC CAT GAC AAT TAC CCA TAC CAC CTC ATT ATG CCC CAT CTC CGC-3′ | 0.01 to 1000 μg/L | 4.8 ng/L | [162] | |
| Chloramphenicol | 5′-AGCAGCACAGAGGTCAGATGACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAGCCTATGCGTGCTACCGTGAA-3′ | 10–107 pg/mL | 0.987 pg/mL | [163] | |
| Electrochemical | OTA | 5′-triple SH-GAT CGG GTG TGG GTG GCG TAA AGG GAG CAT CGG ACA-3′ | 1 × 10−5–10 nM | 3.3 × 10−3 pM | [190] |
| Amoxicillin | 5′-(SH)-TTA GTT GGG GTT CAG TTG G-3′ | 0.5–3 nM | 0.2 nM | [191] | |
| Thrombin | Apt1: 5′-COOH-(CH2)10-GGTTGGTGTGGTTGG-3′. | 1 pM–10 nM | 0.64 pM | [192] | |
| Apt2: 5′-NH2-(CH2)6-AGTCCGTGGTAGGGCAGGTTGGGGTGACT-3′ | |||||
| PSA | 5′-NH2-TTT TTA ATT AAA GCT CGC CAT CAA ATA GCT TT-3′ | 0.0001–100 ng/mL | 0.085 pg/mL | [169] | |
| OTC | 5′-SH-GGA ATT CGC TAG CAC GTT GAC GCT GGT GCC CGG TTG TGG TGC GAG TGT TGT GTG GAT CCG AGC TCC ACG TG-3′ | 1 × 10−13 to 1 × 10−5 g/mL | 3.1 × 10−14 g/mL | [196] | |
| PAT | 5′-NH2-GGCCCGCCAACCCGCATCATCTACACTGATATTTTACCTT-3′ | 5 × 10−8–5 × 10−1 μg/ | 1.46 × 10−8 μg/mL | [43] | |
| Thrombin | 5′-AGTCCGTGGTAGGGCAGGTTGGGGTGACT-3′ | 1 fM–1 nM | 0.57 fM | [13] | |
| Pb2+ | 5′-GGGTGGGTGGGTGGGT-3′ and its complementary strand 5′-CCACCCACCC–(CH2)6–SH-3′ | 0.5–25 ppb | 0.6 ppb | [197] | |
| H5N1 | 5′-biotin-GTGTGCATGGATAGCACGTAACGGTGTAGTAGTAACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCGTGTTG-3′ | 0.001 to 1 HAU | 8 × 10–4 HAU | [201] | |
| Sulfadimethoxine | 5′-GAG GGC AAC GAG TGT TTA TAG A-3′, DNA probe, 5′–SH–TCT ATA AAC ACT CGT TGC CCT C-3′ | 0.1–500 nM | 0.038 nM | [203] | |
| Malathion | 5′-COOH-ATCCGTCACACCTGCTCTTATACACAATTGTTTTTCTCTT AACTTCTTGACTGCTGGTGTTGGCT-3′ | 0.5–600 ng/L | 0.5 ng/L | [204] | |
| ATZ | 5′-TGT-ACC-GTC-TGA-GCG-ATT-CGT-ACG-AAC-GGC-TTT-GTA-CTG-TTT-GCA-CTG-GCG-GAT-TTA-GCC-AGT-CAG-TGT-TAA-GGA-GTG-C-3′ | 50-fM–0.3 nM | 12.0 fM | [206] | |
| OTA | 5′-AAAGATCGGGTGTGGGTGGCGTAA AGGGAGCATCGGACA-3′ | 0.01–1 ng/mL | 0.004 ng/mL | [205] |
Figure 2.
(A) Schematic illustration of AFB1 assay with the label-free colorimetric aptasensor using the competition between the specific aptamer, CPP, and AuNPs inducing AuNP aggregation, reproduced from [90]. (B) Schematic protocol of the dopamine assay via aptamer-assisted AuNP-induced NaCl salt aggregation with two possible mechanisms of sensing, reproduced from [91]. (C) Schemes of the As (III) assay based on the aptamer–As (III) competitive interaction inducing salt–AuNP aggregation, reproduced from [92]. (D) Schematic principle of S. typhimurium assay based on the peroxidase-mimicking activity of dual aptamers@BSA-AuNCs probes inducing a blue color for 3,3′,5,5′-tetramethylbenzidine (TMB), reproduced from [97].
As previously discussed, several colorimetric aptasensors were developed based on AuNP-induced aggregation with different targets such as malathion, Cd2+, Salmonella typhimurium, and AFB1, changing the specific aptamer functionalized with the AuNPs [85,86,87,88,90]. The question is what the role of these targets and others in the stability and instability of AuNPs in solution is, and if this could influence the salt-induced AuNP aggregation or not, affecting the reliability of the proposed strategies.
Recently, Zong et al., and Liu et al. [91,92] proposed a study on the competitive binding affinities of both the aptamer and target toward the AuNPs and the effect of this binding on the sensing strategy based on salt-induced aggregation, as displayed in Figure 2B,C. Liu et al. investigated the adsorption affinity of dopamine as a target and its aptamer on the AuNP surface in an aptamer-based sensing strategy, revealing competition between the aptamer and target on the AuNP surface. To confirm the competition, a proposed competition between two different targets (i.e., melamine and K+) was also evaluated using their specific aptamers. The study aimed to examine if the binding between the target and AuNPs (dopamine and melamine were used as examples for strong target/AuNP interactions, while K+ was used for weak interactions) could affect the analytical reliability of the sensor. Liu et al. claimed two possible mechanisms, in which a aptamer specific for dopamine could adsorb on the surface of AuNP as described in Figure 2(Ba), or the dopamine target had a higher affinity to AuNPs than the aptamer, as shown in Figure 2(Bb). This study reveals that both the aptamer and dopamine have a strong affinity to the AuNP surface, and the dopamine itself induces different degrees of aggregation based on its concentration. A more interesting finding is that the dopamine adsorption on the AuNPs could inhibit the aptamer adsorption, revealing that the most probable mechanism is the one described in Figure 2(Bb). Finally, it was disclosed that the system based on AuNP aggregation-assisting aptamer is strongly kinetically controlled depending on the degree of the binding affinity and competition between the AuNP and both the target and aptamer and, also, depending on whether this target can also induce the stabilization or destabilization of AuNPs [91]. Based on this study, the free target in solution could cause problems for the sensor reliability, as also proposed by Zong et al. [92] in their study on the assay of As+3, as shown in Figure 2C. Therefore, the use of a stronger capping agent on AuNPs (cationic AuNPs or coating with nanomaterials such as graphene) that cannot be displaced compared to citrate could decrease the target’s strength of adsorption to the AuNP surface and avoid these limitations [93,94,95,96].
The AuNPs were confined for using in a colorimetric aptasensor with different strategies other than aggregation, depending on the catalytic performance of the NPs and anisotropic growth of the AuNPs. Chen et al. proposed a colorimetric aptasensor for the selective and sensitive detection of Salmonella typhimurium (S. typhimurium) based on S. typhimurium’s affinity, to enhance the peroxidase-like activity of bovine serum albumin-stabilized gold nanoclusters modified with dual aptamers (aptamers@BSA-AuNCs), as outlined in Figure 2D [97]. In the presence of S. typhimurium, the aptamers@BSA-AuNCs could oxidize colorless 3,3′,5,5′-tetramethylbenzidine (TMB), producing the blue oxidized form, with a wide linear response to S. typhimurium in the concentration range of 101–106 CFU/mL and LOD of 1 CFU/mL [97].
Wang et al. proposed a colorimetric strategy for abscisic acid (ABA) detection based on the anisotropic growth of AuNPs in the presence of aptamer on the surface of the seed AuNPs [98]. In the presence of ABA, the functionalized aptamer on the surface of the AuNPs changed into a G-quadruplex-like structure and bound with ABA, triggering the departure of the aptamer from the surface of the AuNPs. In the presence of both the growth-promoting HAuCl4 and NH2OH as reducing agents, a uniform or anisotropic growth of AuNPs with a distinct change in the localized surface plasmon resonance (LSPR) could occur based on the amount of bound aptamer on the surface of the bare seed AuNPs, which is related to the concentration of ABA, with a linear relationship for 1 nM to 10 μM and LOD of 0.51 nM.
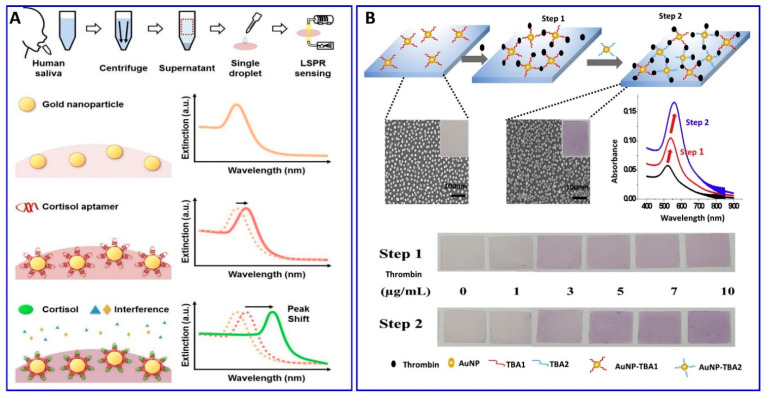
Furthermore, Seongjae Jo et al. and Wonjung Lee et al. [99,100] described a fabricated solid aptamer sensor for assaying cortisol and thrombin, respectively, by immobilizing the a specific designated aptamer with a specific sequence outlined in the (Table 1) on a glass substrate, as shown in Figure 3A,B. The changes in the LSPR, in the presence of cortisol and thrombin, showed linear ranges of 0.1–1000 nM and 0–10 μg/mL, with LODs of 0.1 nM and 1.33 μg/mL, respectively.
Figure 3.
(A) Schematic illustration of the cortisol assay based on localized surface plasmon resonance (LSPR) aptasensor, reproduced from [99]. (B) Scheme representing the thrombin assay via sandwich colorimetric solid-phase aptasensor based on the enhanced LSPR, reproduced from [100].
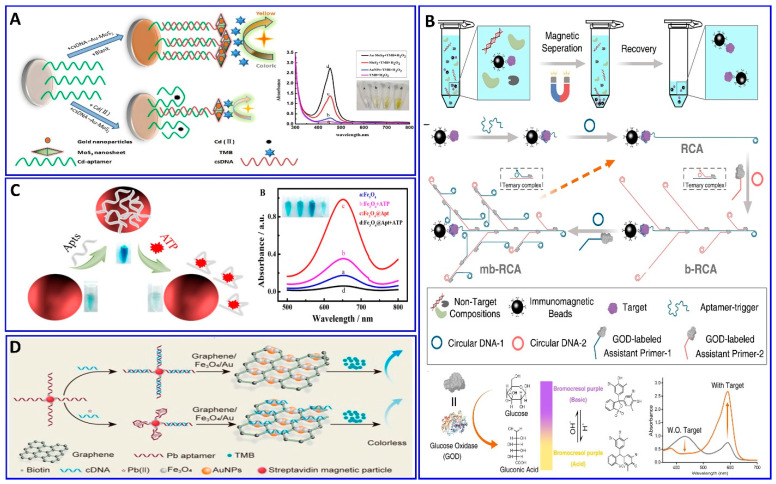
A colorimetric strategy other than AuNP aggregation was developed for a Cd2+ assay assisted by a specific Cd2+ aptamer based on the enzyme-mimicking activity of MoS2 nanocomposites, as reported by Tao et al. [101] (Figure 4A). The biotinylated Cd2+ aptamer was immobilized on the bottom of the microplate via biotin–avidin affinity; simultaneously, a functionalized complementary strand CsDNA–Au–MoS2 was used as a signal transducer, depending on its peroxidase-like activity toward the chromogenic TMB. This system exhibited a linear range of 1–500 ng/mL and a LOD of 0.7 ng/mL.
Figure 4.
(A) Principal strategy for Cd2+ assay via the enhanced peroxidase-like activity of Au–MoS2-based aptamer, reproduced from [101]. (B) Principal protocol for the PDGF-BB assay via a glucose oxidase enzyme, inducing changes in pH-equipped immunomagnetic beads and mb-RCA as a signal amplifier, reproduced from [103]. (C) Schematic diagram showing the procedure and mechanism of the ATP assay via colorimetric aptamer technology based on the peroxidase-like catalysis properties of Fe3O4, reproduced from [106]. (D) Colorimetric procedure for Pb2+ assay based on the peroxidase-mimicking activity of graphene/Fe3O4-AuNPs aptamer technology, reproduced from [107].
Magnetic nanoparticles (MNPs) have also been used as a nanoassembling template for bioassay targeting, either alone or in combination with other nanostructures, where they can be easily isolated from the solution via an external magnetic field [39,102,103,104,105]. Miao et al. reported a sensitive pH-based colorimetric strategy based on glucose oxidase (GOD) enrichment and catalysis for the amplified detection of cancer biomarkers with isothermal DNA signal amplification and multibranched rolling circle amplification (mb-RCA) [103]. The mb-RCA is integrated with a magnetic separation technology to avoid potential interference, as shown in Figure 4B; the system successfully detected the carcinogenic human PDGF-BB biomarker with a LOD of 0.94 pM [103]. The Fe3O4 nanoparticles incorporated with aptamers were used for assaying trace levels of adenosine triphosphate (ATP) biomarkers colorimetrically, as provided by Li et al. in Figure 4C [106]. The catalytic activity of the Fe3O4 nanoparticles in the conversion of the colorless TMB into blue TMB was enhanced significantly after functionalization with an ATP aptamer. In the presence of an ATP target, the target binds with its aptamer, which departs from the surface of the Fe3O4, decreasing its activity, with a detection range of 0.50–100 μM and a LOD of 0.09 μM. The toxic Pb2+ was also assayed via a colorimetric aptasensor based on the peroxidase-like activity of graphene/Fe3O4-AuNP composites, showing a linear range of 1–300 ng/mL and a LOD of 0.63 ng/mL, as reported by Tao et al. [107] (Figure 4D).
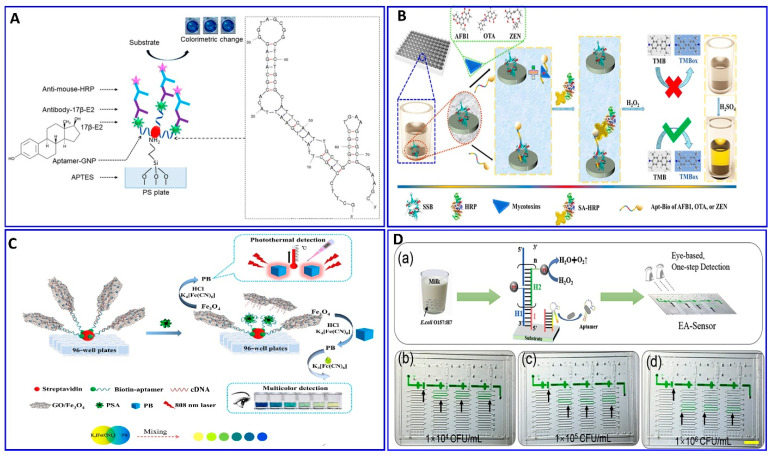
Replacing the antibodies with aptamers in the ELISA protocol has attracted attention for assaying food-borne pathogens (e.g., E. coli), as reported by Duan et al.’s group [108]. First, the biotin aptamer was immobilized on the surface of an avidin-coated microplate to capture the E. coli bacteria. The detection aptamer was fabricated with Cu-MOF NPs through avidin–biotin affinity as a transducer signaling based on the peroxidase activity of the Cu-MOF NPs catalyzing the conversion of the colorless TMB into the oxidized blue structure and, then, yellow in the presence of the acid. This modified ELISA succeeded in detecting 16 to 1.6 × 106 CFU/mL, with a LOD of 2 CFU/mL [108]. The introduction of the aptamer technology in the sandwich ELISA was used in the detection of 17β-estradiol-E2 (17β-E2) by Huang et al. [109] (Figure 5A). Xing et al. reported the detection of toxins, AFB1, ochratoxin A, and zearalenone, with a green ELISA based on a single-stranded-binding protein (SSB)-assisted aptamer with horseradish peroxidase (HRP) technology to produce the signal as demonstrated in Figure 5B [110]. This modified ELISA aptamer showed low limits of detection in corn: 112, 319, and 377 ng/L for AFB1, ochratoxin A, and zearalenone, respectively. Wei et al. described a multicolor and photothermal assay of prostate-specific antigen (PSA) as a dual signals based on ELISA modified aptamers, as outlined in Figure 5C [111]. In the modified ELISA, an Fe3O4 NP–graphene oxide composite was functionalized with complementary DNA and served as the signal producer for assaying the PSA. In the absence of PSA, the Fe3O4 NP–graphene oxide–DNA probes were captured by the PSA aptamer; therefore, the Fe3O4 NPs were transformed into Prussian blue (PB) using a HCl solution, which was then mixed with potassium ferricyanide to produce a multicolor signal; simultaneously, the PB can produce a thermal signal upon near-infrared laser irradiation, with LODs of 0.15 and 0.31 ng/mL for colorimetric and photothermal assays.
Figure 5.
(A) Schematic illustration of the ELISA for assaying of 17β-E2 with an assisting aptamer–antibody sandwich functionalized with AuNPs, reproduced from [109]. (B) Principal assay procedures for mycotoxins using a green ELISA based on a single-stranded binding protein (SSB)-assisted aptamer; the SSB was dispensed into polystyrene 96 plates; reproduced from [110]. (C) Schematic illustration outlining the multicolor and photothermal assay for prostate-specific antigen (PSA) via magnetic beads assisting aptamer separation, reproduced from [111]. (D) Detection of E.coli O157:H7 in milk via the developed naked-eye EA-Sensor, reproduced from [112].
Li et al. introduced a simple, eye-based microfluidic aptasensor (EA-Sensor) for the rapid assay of E.coli O157:H7 [112], as described in Figure 5D. Coupling between the aptamer and hybridization chain reaction (HCR) amplification was considered based on distance visualized readout technology for quantitative assays (Figure 5D). The HCR amplified the signal 100-fold after the E.coli O157:H7 was recognized by the aptamers. The system exhibited good linearity in the range of 500–5 × 107 CFU/mL and a LOD of 250 CFU/mL.
3. Fluorescence Aptasensor
Fluorescence-based aptasensors are characterized by high sensitivity, large-detection ranges, multiplexing capabilities, rapid assaying, and the highly selective recognition of aptamers for several targets, which can be distinguished under UV light, in comparison with the colorimetric sensor’s signals that are distinguished using visible light. The integration of both fluorescent materials (fluorophore dyes and fluorescent nanoparticles such as upconversion nanoparticles (UCNPs), GO, and CQDs) and aptamers can produce high sensitivity and selectivity, and a rapid analysis strategy, making them useful candidates for fluorescence-aptasensor bioassays [113,114,115]. The recognition affinity between aptamers and analytes induces conformational changes in the aptamer. This process can trigger changes in the fluorescent emission properties of the fluorophore dye or fluorescent nanomaterials, owing to changes in the original environments of these materials.
The design of fluorescent aptasensors requires the use of hairpin aptamers (aptabeacons), which are labeled with either a fluorophore or a quencher. Forster resonance energy transfer (FRET) typically uses a donor fluorophore and an acceptor quencher material. Different strategies are often employed via FRET operations, which are performed through either “signal-on” or “signal-off” assaying protocols. These operations are based on the disparity in the fluorescence responses of the fluorophores as a function of the potential and unique aptamer–target binding and the conformational-change degree [116,117,118]. Owing to the conformational change of the aptamer induced via target interactions, the probe was successfully switched on/off via the FRET mechanism.
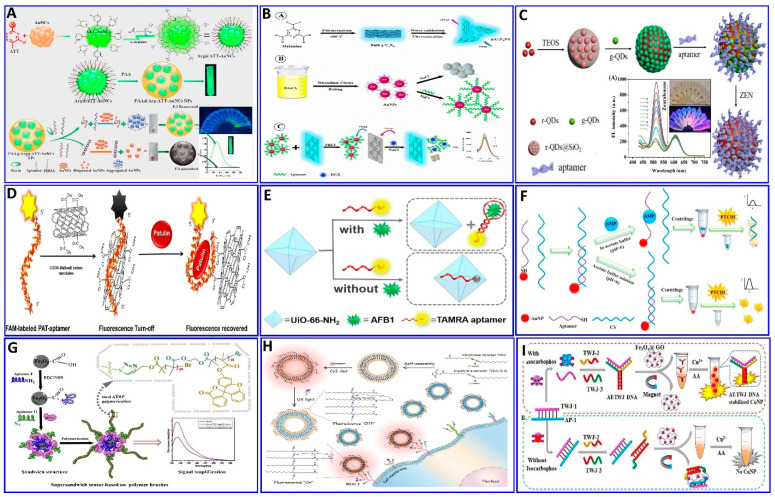
Some of the relevant fluorescent strategies were employed for the assaying of several targets based on integrated aptamers and fluorescent materials as described in the following Figure 6, Figure 7 and Figure 8. Khan et al. provided a colorimetric and fluorometric method for the assaying of T-2 (trichothecenes A) with the assistance of aptamers [119], as indicated in Figure 6A. In this method, through the FRET mechanism, a green-emitting L-arginine@6-aza-2-thiothymine-protected gold nanocluster capped with polyacrylic acid (PAA@Arg@ATT-AuNCs) was quenched via dispersed AuNPs in the absence of the T-2 target. In the presence of the target, the PDDA became free in the solution and induced the aggregation of the AuNPs that were unable to quench the fluorescence probe (LOD: 0.57 pg/mL, and linear range: 0.001−100 ng/mL) [119].
Figure 6.
(A) Schematics showing the preparation of PAA@Arg@ATT-AuNC as a fluorescent probe for trichothecenes A (T-2) toxin assay, reproduced from [119]. (B) Graphical illustration for the assaying of digoxin (DGX) using the aptamer/AuNPs/g-C3N4NS sensor probe, reproduced from [120]. (C) Schematic illustration of the zearalenone assay using a ratiometric fluorescent nanoprobe with dual emission at 518 and 608 nm, reproduced from [121]. (D) Schematic description of the protocol employed for a patulin (PAT) assay via CA-MWCNTs) with quenching of aptamer-tagged carboxyfluorescein, reproduced from [130]. (E) Schematic description of AFB1 detection based on UiO-66-NH2 and aptamer-functionalized TAMRA dye, reproduced from [123]. (F) Schematic of the fluorescent aptasensor for an AMP assay based on the competitive quenching between the AMP aptamer complementary strand and AuNPs, reproduced from [131]. (G) Schematic showing an ultrasensitive fluorometric aptasensor for IFN-γ detection by dual atom transfer radical polymerization (ATRP) amplification, reproduced from [132]. (H) Schematic showing the fabrication of PDA–Apt liposomes and fluorescence imaging of plasma membrane glycoprotein MUC1, reproduced from [133]. (I) Schematic description of isocarbophos assay via fluorometric aptsensor based on AT-rich three-way junction DNA template copper nanoparticles and Fe3O4@GO, reproduced from [134].
Figure 7.
Schematic description of the (A) sensing platform for E. coli detection using fluorescent UCNP–WS2 nanosheet, reproduced from [140]. (B) Proposed fluorescence aptasensor for E. coli. using MNP–aptamer–cDNA–UCNP probe, reproduced from [141]. (C) Fluorometric aptasensor for malathion assay based on the FRET between UCNPs and GNPs, reproduced from [142]. (D) Proposed scheme of ATP assay based on ratiometric fluorescence from the binding between the ATP and aptamer complexes, reproduced from [145].
Figure 8.
(A) Schematic illustration of the ATP assay via quenching of DNA/AgNCs by AuNRs functionalized with aptamer and Exo III for signal amplification, reproduced from [152]. (B) Fluorometric assay for acetamiprid using exonuclease integrated with the aptamer protocol, reproduced from [154]. (C) Exosomal assay based on the AIE mechanism using graphene oxide (GO) as a quencher for tertiary amine-containing tetraphenylethene (TPE-TA) dye, reproduced from [121]. (D) Dual-mode fluorescent aptasensor using both aptamer and a DNAzyme to assay ATP with two different mechanisms, fluorometric and colorimetric signals, reproduced from [159]. (E) TTX assay based on the difference in fluorescence response of the berberine reporter, reproduced from [160]. (F) Turn-on fluorescence aptasensor assay for chloramphenicol based on oligomer quencher release, reproduced from [163].
Some studies have focused on detecting several targets based on the aptamer-competitive quenching of carbon quantum dots and graphene materials [120,121,122,123,124,125]. Recently, GO was used as a “nanoquencher” for fluorescent materials via the FRET strategy with ssDNA as a nanoscaffold, capable of adsorbing to the GO surface through π–π stacking interactions with the nucleotide bases [126,127,128,129]. Shirania et al. proposed a fluorometric aptasensor for assaying digoxin (DGX) based on the quenching effect of an aptamer/gold nanoparticle probe on the fluorescence intensity of graphitic carbon nitride nanosheets. The FRET strategy was employed, and the presence of DGX resulted in fluorescence intensity restoration (Figure 6B), with concentrations ranging from 10 to 500 ng/L and a LOD of 3.2 ng/L [120]. Tan et al. [121] proposed another protocol system that uses a ratiometric fluorescent nanoprobe, in which dual emission at 518 and 608 nm is induced by a single excitation. This system was applied for the assaying of zearalenone (ZEN), which could quench green-emitting g-QDs linked to an aptamer via an electron transfer mechanism (Figure 6C), with a LOD of 7.5 nM [121]. Khan et al. proposed that the water-soluble mycotoxin patulin (PAT) produced by several fungal species, including from the genera Aspergillus, Penicillium, and Byssochlamys, be assayed via fluorometric aptasensors (Figure 6D), where the carboxyfluorescein (CFL) fluorescent dye tagged with a PAT aptamer was quenched by –COOH-functionalized multiwall carbon nanotubes (CA-MWCNTs). The presence of PAT induces the recovery of the fluorescence, with a LOD of 0.13 ng/mL [130].
Recently, Guo et al. reported that thrombin and ATP aptamers exhibited quenching effects on positively prepared polyethylenimine (PEI-CDs). In the presence of thrombin or ATP, the respective aptamer binds with its target, leading to the restoration of the fluorescence, with LODs of 1.2 and 13 nM for thrombin and ATP, respectively [95]. The aptamer has been functionalized with fluorescent dyes such as the TAMRA dye, as proposed for the assaying of AFB1 (Figure 6E) [123]. In the absence of AFB1, the specific aptamer labeled with TAMRA was adsorbed on the surface of the UiO-66-NH2 (metal–organic framework, MOF). This MOF can quench the fluorescence of the TAMRA with a sensitivity of 0–180 ng/mL and a LOD of 0.35 ng/mL. 3,4,9,10-perylenetetracarboxylic acid diimide (PTCDI), an affordable and low-cost fluorophore dye, was used for the assaying of ampicillin (AMP). This assaying was based on the competitive quenching between the AMP aptamer, and the complementary strand (CS) and AuNPs (Figure 6F), with a linear range of 100 to 1000 pM and LOD of 29.2 pM [131].
A method for signal amplification was introduced by Wen et al., who applied dual atom transfer radical polymerization (ATRP) technology for amplifying the fluorogenic signal during the assaying of gamma-interferon protein (IFN-γ). As outlined in Figure 6G [132], two aptamers were used to sandwich the protein, where the second aptamer was functionalized with azide, which, upon polymerization, produces an amplified fluorogenic signal (LOD: 0.178 fM).
Polydiacetylene (PDA) liposomes were immobilized with an aptamer-tagged fluorophore (Cy3–Apt) for assaying mucin 1 (MUC1), as indicated in the proposed scheme shown in Figure 6H, with a LOD of 0.8 nM [133], based on the spectral overlap of Cy3’s fluorescence emission and the absorbance of PDA–Apt liposomes [133]. Fan et al. synthesized Trich three-way junction (AT-TWJ) DNA-stabilized CuNPs as a fluorescent transducer and magnetic Fe3O4@GO as a single-stranded DNA (ssDNA) adsorbent for the detection of isocarbophos [134] (Figure 6I). The dsDNA is hybridized by the isocarbophos aptamer, while the complementary DNA plays a role as a recognition unit. In the presence of isocarbophos, the released dsDNA and the two ssDNA strands can assemble into AT-TWJ DNA. This facilitates the formation of AT-TWJ DNA template CuNPs with a strong fluorescence response toward isocarbophos (concentration range: 10–500 nM), with a LOD of 3.38 nM.
Carbon nanotubes (CNTs) are considered perfect nanoquenchers for fluorophore-labeled ssDNA, owing to the stacking of nucleotides on the surface of the multiwalled carbon nanotubes (MWCNTs), which induces quenching via the FRET system. The hybridization of the aptamer with the target leads to the restoration of the fluorescence. Therefore, the ssDNA–MWCNT assembled fluorescence probe was used for the assaying of Hg2+ [135].
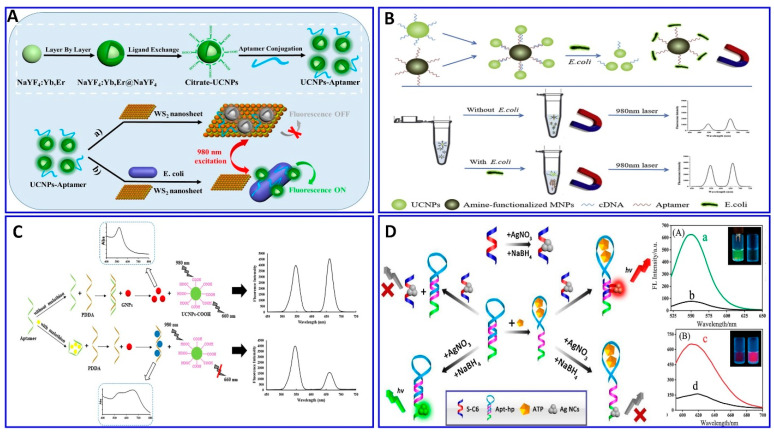
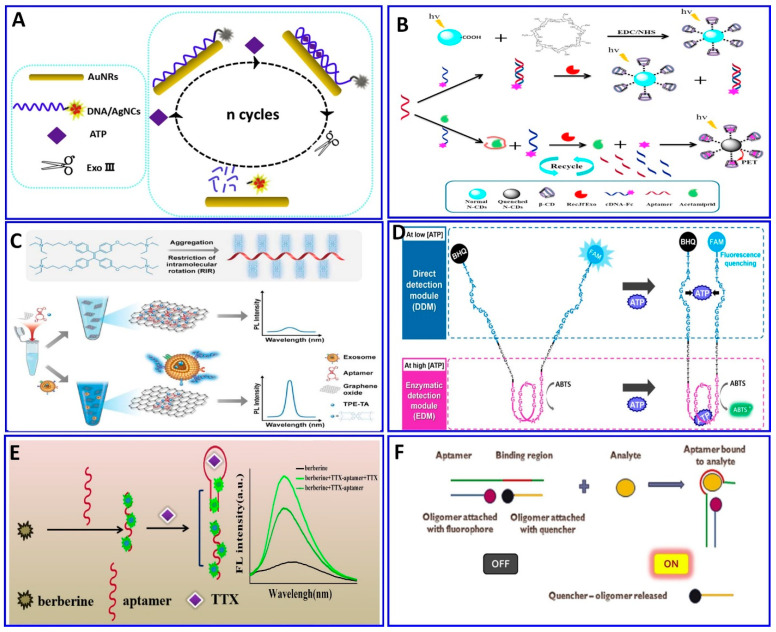
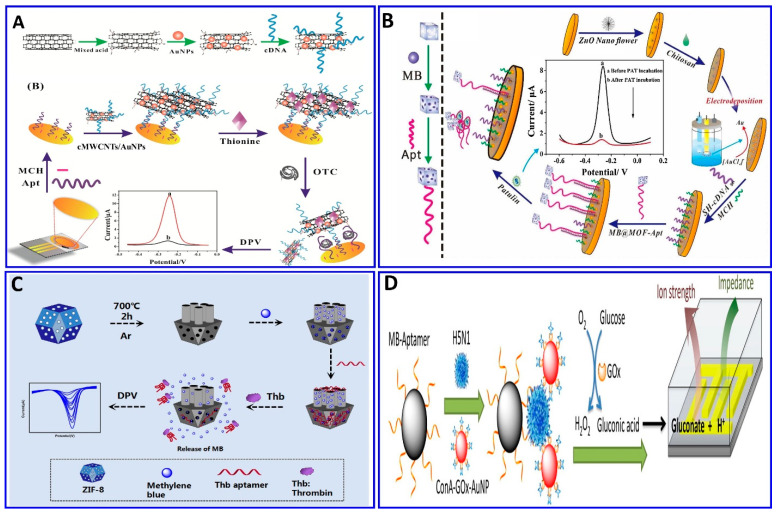
Rare-earth-element-doped UCNPs can convert low-energy light into high-energy light with short wavelengths via the photon mechanism. UCNP materials are characterized by high quantum yields, narrow emission peaks, excellent photostability, long fluorescence lifetimes, environmental friendliness, low toxicity, bulky anti-Stokes shifts, and high resistance to photobleaching. Moreover, infrared light induces minimal photodamage to the biological targets without exciting the biological fluorophoric substance, which can be confined in the matrix [80,136,137,138,139]. UCNPs were used in the development of fluorescence aptasensors for assaying different targets [140,141,142,143,144]. However, they still have some limitations such as the difficulty in distinguishing different target concentrations by the naked eye upon excitation by a laser beam at 980 nm, where the target can be only be detected using instrumentation. However, UCNPs have been used for aptasensing development, in which a new biosensor for E. coli bacteria has been developed based on the FRET between aptamer-functionalized UCNPs as donors and layered tungsten disulfide (WS2) as the acceptor. This was achieved through the attachment of the aptamer to the WS2 surface via van der Waals forces. In the presence of E. coli, the specific aptamer binds preferentially to E. coli, causing a conformation of the aptamer that induces the dissociation of the probe away from the surface of the WS2 nanosheets. Therefore, part of the fluorescent intensity is restored as a function of E. coli (Figure 7A), with concentrations ranging from 85 to 85 × 107 CFU/mL and a LOD of 17 CFU/mL [140]. Moreover, E. coli was detected using immobilization between UCNPs and aptamer technology as described in Figure 7B, in which the fluorescence emission intensity of the MNP–aptamer–cDNA–UCNP complex decreases upon increasing the E. coli concentration, with a linear system range of 58–58 × 106 CFU/mL and LOD of 10 CFU/mL [141]. A similar protocol was employed for a malathion assay using UCNPs (Figure 7C), with a linear range of 0.01 to 1 μM and LOD of 1.42 nM [142]. Pb2+ was detected using aptamer-functionalized UCNPs incorporated with magnetic Fe3O4− (MNPs), with modified AuNPs, with a detection range of 25–1400 nM and LOD of 5.7 nM [143]. Basically, the dispersion and aggregation affinity were harnessed to produce a quencher for the fluorescent materials, as used in the assaying of chlorpyrifos (CPF) [144]. Fluorescent terbium (III)-based MOFs (Tb-MOFs) were quenched by dispersed AuNPs, and the fluorescent intensity was restored by PDDA-aggregated AuNPs, owing to the binding between the aptamer and CPF, with a sensitivity of 5–600 nM and LOD of 3.8 nM [144]. Qiu et al. proposed a ratiometric biosensing fluorescent sensor for ATP based on multicolor silver nanoclusters (AgNCs; Figure 7D) [145]. Once the sensor is exposed to the ATP, the binding between the ATP and aptamer complexes opens the hairpin structure and releases the anchor sequence, which hybridizes with the ssDNA and induces the turning off of the green fluorescence, while the red fluorescence is turned on, with a LOD of 0.38 μM [145].
As mentioned above, aptamers are sequences of nucleic acid, and because of that, they can be integrated with a DNA system for fluorescent signal amplification to improve the detection limit and, therefore, be used in the early diagnosis of diseases. These amplification processes include rolling-cycle amplification, strand displacement reactions, hybridization chain reactions, and hairpin DNA cascade hybridization reactions [146,147,148,149,150,151]. Ning et al. used exonuclease III (Exo III)-assisted target-recycling amplification integrated with aptamer technology for the ultrasensitive assaying of adenosine triphosphate (ATP) via the quenching of the DNA/AgNCs fluorescent probe with gold nanorods (+) (AuNRs), with a LOD of 26 pM [152], as described in Figure 8A. The amplification of the signal using exonuclease I (Exo I) was also integrated with the fluorescent aptasensor for assaying AFB1 [153]. In the presence of AFB1, the aptamer structure changes, preventing its digestion by Exo I. Therefore, the addition of SYBR intensifies the fluorescence as a function of the AFB1 concentration, with a wide linear range of 2–400 ng/mL and LOD of 1.82 ng/mL [153]. Employing the same technology, the exonuclease integrated with an aptamer was used to assay acetamiprid [154]. In the absence of acetamiprid, the specific aptamer hybridizes with the complementary DNA labeled with ferrocene (cDNA–Fc), without a change in the fluorescence intensity after the addition of the exonuclease (RecJf), while in presence of acetamiprid, it will bind with the aptamer, which leaves the cDNA–Fc, and then digested by the RecJf exonuclease, liberating the Fc to interact with cyclodextrin and initiating photoinduced electron transfer, as outlined in Figure 8B [154].
Aggregation-induced emission (AIE) was conducted for assaying exosomal tumor-associated proteins using GO as a quencher and TPE-TA (tertiary amine-containing tetraphenylethene) as a fluorescent dye (Figure 8C). The positively charged TPE-TA binds one aptamer and aggregates, resulting in an amplified fluorescence. When the target exosomes are introduced, the aptamer preferentially binds with its target. Thus, the TPE-TA/aptamer complexes detach from the GO surface, inducing the “turning on” of the fluorescence, with a response range of 0.68–30.4 pM and a LOD of 0.57 pM [155].
The aptamers themselves can be modified with a fluorophore and quencher materials at both ends without affecting their binding affinity toward the targets, so a FRET aptasensor strategy can be easily constructed in the presence of targets inducing fluorescence quenching [156,157,158]. Kang et al. provided a dual-mode readout DNA biosensor combining both an aptamer and a DNAzyme to assay ATP with two different mechanisms, using fluorometric and colorimetric signals (Figure 8D), respectively. The presence of ATP induces attraction between the two strands of the aptamer, which induces the quenching of the fluorescence dye on one aptamer end via the quencher labeled on the other end of the aptamer. At the same time, the ATP induces the folding of the DNAzyme into a G-quadruplex, enhancing its peroxidase activity in oxidizing 2′-azino-bis(3-ethylbenzothiazoline-6- sulfonic acid) (ABTS). The system showed a large dynamic range, 1−500,000 μM, more than five orders larger than that reported to date for ATP assays [159]. Lan et al. used a strategy for assaying tetrodotoxin (TTX) based on the difference in the fluorescence response of berberine as a result of TTX–aptamer interaction with the analyte (Figure 8E), exhibiting a linear range of 0.1–500 nM and a LOD of 0.074 nM [160]. A molecular imprinting polymer (MIP) was used for assaying Cd2+ in environmental and agricultural samples with the assistance of a specific aptamer. The MIP was integrated with aptamers to make dual recognition units, while carbon-doped sulfur, nitrogen quantum dots, and AuNPs (SN-CQD/Au) were used as a fluorophore. The fluorescence was quenched via Cd2+, with a linear response range of 20 pM–12 nM and a LOD of 1.2 pM. [161].
Zhang et al. developed a simple, sensitive, and label-free method to assay the cyanotoxin microcystin–leucine–arginine (MC-LR), using fluorescent dsDNA–CuNCs as a fluorescent probe, comprising an aptamer for MC-LR and its complementary ssDNA. The presence of the MC-LR target results in the uptake of the aptamer from the dsDNA–CuNCs; therefore, the fluorescent signal exhibits a sensitivity range of 0.01 to 1000 μg/L and LOD of 4.8 ng/L [162]. Sharma et al. provided a technique for chloramphenicol analyte detection using three oligonucleotide complexes comprising an aptamer, fluorophore oligomer, and oligomer quencher as a sensor platform (Figure 8F), with a linear range of 10–107 pg/mL and a LOD of 0.987 pg/mL [163].
4. Electrochemical Aptasensor
An electrochemical aptasensor was fabricated using an aptamer as a bioreceptor and an electrochemical transducer, which translated the target–aptamer affinity into a measurable electrochemical signal through potentiometry, voltammetry, amperometry, impedimetry, or electrochemiluminescence. The potentiometric approach involves the measurement of the potential between the probe and the reference electrode without any net charge transfer. The amperometric approach is based on applying a potential and allowing a redox reaction to occur. The signal is defined as the current between the electrode and the counter-electrode. The voltammetric strategy includes the sweeping potential over time and recording the corresponding current; this strategy is based on a three-electrode system, with working, reference, and counter electrodes. The impedimetric technique involves measuring the charge transfer rate on the surface of the electrode for a kinetic study.
The electrochemical aptasensors were modified with various nanomaterials such as carbon-based nanomaterials, metal–organic frameworks (MOFs), AuNPs, and polymers for signal amplification [164,165,166,167]. The aptamers were immobilized on the electrode surface via π–π interactions, hydrogel grafts, biotin–avidin interactions, thiol–gold self-assembly, and carboxyl–amine covalent reactions [168,169,170,171,172,173].
The electrochemical aptasensor mainly depends on the interactions occurring on the surface of transducer as a result of the induced reaction between the target and its specific aptamer, providing amperometric or potentiometric electrochemical signals. Another technique is based on the increase in charge transfer resistance via the impedance technique [174]. Therefore, an electrochemical aptasensor was provided for the detection of several targets, such as ampicillin (AMP), avian influenza virus (H5N1), carbohydrate antigen 125, Pb2+, lysozymes, insulin, thrombin, CD44, vanillin, circulating human MDA-MB-231 breast cancer cells, bisphenol A, furaneol, and Hg2+, among others [8,46,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189].
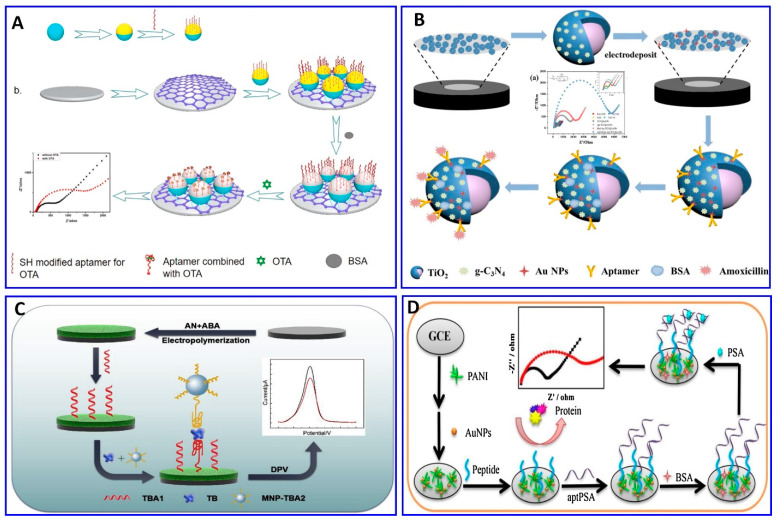
The chratoxin A (OTA) toxin was detected electrochemically, specifically using the OTA aptamer as demonstrated in Figure 9A. The probe was fabricated via the immobilization of the OTA aptamer on the gold layer, and the surface of the aminated polystyrene, which binds with the glassy carbon electrode (GCE) via peptide bonds, was coated with the aptamer functionalized with gold, exhibiting a sensitivity of 1 × 10−5 to 10 nM, and a LOD of 3.3 × 10−3 pM [190]. Amoxicillin was detected in wastewater via an ultrasensitive impedimetric aptasensor based on the synergetic effect of the surface of the GCE being coated with TiO2-g-C3N4 and AuNPs (Figure 9B). Because of the formation of the Au–S bond between the AuNPs and the modified thiol aptamer, competition on the surface of the GCE occurs between the amoxicillin and its aptamer, achieving a detection sensitivity of 0.5–3 nM and a LOD of 0.2 nM [191].
Figure 9.
(A–D) Schematic illustrations for the fabrication of electrochemical aptasensors. (A) Impedimetric assay using glassy carbon electrode (GCE) immobilized with OTA aptamer, reproduced from [190]. (B) Impedimetric assay for amoxicillin based on the synergetic effect between the amoxicillin and its aptamer on the surface of the GCE coated with TiO2-g-C3N4-AuNPs, reproduced from [191]. (C) A sandwich-type aptasensor for thrombin assay using CSPH hydrogel-modified electrode, reproduced from [192]. (D) PSA assay based on polyaniline (PANI)/AuNPs as a conducting layer on GCE immobilized with a PSA aptamer, reproduced from [193].
A sandwich-type electrochemical aptasensor has been fabricated for a thrombin assay, as demonstrated in Figure 9C. The sensor probe was fabricated on a conductive supramolecular polymer hydrogel (CSPH) modified electrode, on which the thrombin-binding aptamer 1 (TBA1) was immobilized via amide bonds, while the thrombin-binding aptamer 2 was modified using magnetic nanoparticles (MNP-TBA2) as signal amplification probes; the sandwich-type electrochemical aptasensor showed a linear range of 1 pM to 10 nM, with a LOD of 0.64 pM [192]. A sensitive electrochemical aptasensor for detecting prostate-specific antigen (PSA), based on coral-like polyaniline (PANI)/AuNPs and having peptides as antifouling materials to reduce nonspecific adsorption, was fabricated on a GCE surface (Figure 9D). A PSA aptamer was immobilized on the electrode for a PSA assay, with a wide linear range from 0.1 pg/mL to 100 ng/mL and a LOD of 0.085 pg/mL [169].
Recently, enzymes such as HRP, glucose oxidase, and alkaline phosphatase, and electroactive compounds such as QDs, ferrocene (Fc), ferrocyanide, methylene blue (MB), and Cd nanoparticles were successfully incorporated in electrochemical aptasensor technologies to be used as signal enhancers [114,194]. A different strategy was integrated in an electrochemical aptasensor for assaying different targets—AFB1-activated protein C, OTC, patulin (PAT), thrombin, and Pb2+, among others—by monitoring the electrochemical signals of the released labeling substrates as a result of competitive targets [13,43,195,196,197,198,199,200]. A portable electrochemical aptasensor was fabricated for assaying OTC using a AuNP/cMWCNT/cDNA@thionine probe as a signal-releasing tag in the presence of the target [196], as outlined in Figure 10A. In the presence of OTC, the AuNP/cMWCNT/cDNA@thionine tag was released from the surface of the gold electrode, inducing a change in the electrical signal, with a linear range of 1 × 10−13–1 × 10−5 g/mL and a LOD of 3.1 × 10−14 g/mL. The same technology was also used for assaying PAT; MOF@Methylene blue was used as the signal tag released from the electrode surface as a function of the PAT concentration, with a linear range of 5 × 10−8–5 × 10−1 μg/mL and a LOD of 1.46 × 10−8 μg/mL [43], as shown in Figure 10B. A homogeneous electrochemical aptasensor was fabricated based on the departure of a target-responsive label, the electroactive dye methylene blue (MB), from an aptamer-gated zeolitic imidazolate framework-8 (ZIF-8) as a function of the thrombin concentration (Figure 10C), with a linear range of 1 fM to 1 nM and LOD of 0.57 fM [13]. Employing the same technology, Pb2+ was detected with a range of 0.5–25 ppb and a LOD of 0.6 ppb, using toluidine blue as a redox-released tag signal liberated in the presence of Pb2+ [197]. Enzymatic catalysis was incorporated in an electrochemical aptasensor for signal amplification [201,202]. Fu et al. assayed H5N1 using the electrochemical aptasensor method integrated with enzymatic catalysis, as shown in Figure 10D [201]. Magnetic beads were fabricated with an H5N1 aptamer to enhance the selectivity, followed by immobilization with concanavalin A (ConA), GOx, and AuNPs for signal enhancement in the glucose-fluid-inducing formation of gluconic acid. The system showed a LOD of 8 × 10–4 HAU (hemagglutination units) in a 200 μL sample [201].
Figure 10.
(A–C) Schematic protocols for electrochemical aptasensor based on the target-responsive label electroactive dye amplification strategy. (A) OTC assay using AuNP/cMWCNT/cDNA@thionine probe as a signaler releasing thionine, reproduced from [196]. (B) PAT assay using MOF@Methylene blue signal tags, with signals as a function of PAT concentrations, reproduced from [43]. (C) Thrombin assay based on target-responsive methylene blue (MB) release from the ZIF-8 surface as a function of the thrombin concentration, reproduced from [13]. (D) Schematic protocols for an electrochemical aptasensor based on an enzymatic catalytic reaction, reproduced from [201].
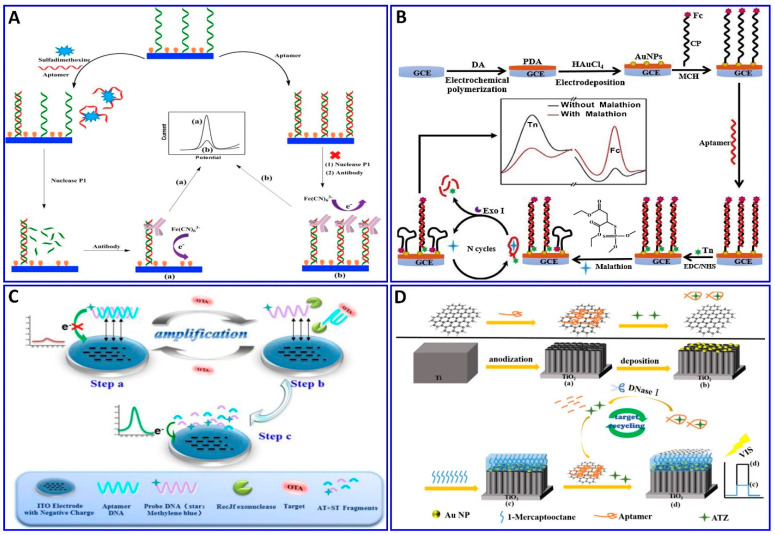
The electrochemical method was integrated with amplification strategies for assaying numerous targets, such as sulfadimethoxine, malathion, OTA, kanamycin, ampicillin, atrazine (ATZ), and acetamiprid [138,203,204,205,206,207,208,209,210]. Sulfadimethoxine was assessed electrochemically with the assistance of nuclease amplification (Figure 11A), in which a gold surface of an electrode was immobilized with a DNA probe, and then hybridized with a specific aptamer for sulfadimethoxine, forming a dsDNA that inhibited their digestion via nuclease P1. The sulfadimethoxine aptamer leaves the DNA probe in the dsDNA in the presence of sulfadimethoxine, allowing the digestion of the dsDNA via nuclease P1 and inducing the amplification of the electrochemical signal, with a response range of 0.1–500 nM and a LOD of 0.038 nM [203]. Malathion was detected with the assistance of exonuclease I (Exo I) as an electrochemical signal enhancer, which enhanced the signal by over two times, as shown in Figure 11B. Moreover, the electrodeposition synthesis of PDA–AuNPs provided satisfactory biocompatibility and electrical conductivity for the sensor. Exo I was used to promote the autocatalytic cycling of malathion by enhancing the current change, with a linear response range of 0.5–600 ng/L [204]. OTA and ATZ were detected electrochemically and photoelectrochemically with the assistance of cycling amplification, with a linear range of 0.01 to 1 ng/mL and a LOD of 0.004 ng/mL for OTA [205] and linear range of 50 fM to 0.3 nM with LOD of 12.0 fM for ATZ [206], as outlined in Figure 11C,D.
Figure 11.
(A–C) Schematic protocols for electrochemical aptasensor integrated with signal amplification based on nuclease strategy. (A) Assaying of sulfadimethoxine using gold electrode fabricated with dsDNA consisting of DNA probe and specific aptamer for sulfadimethoxine inhibiting nuclease digestion, reproduced from [203]. (B) Malathion assay with the assistance of Exo I as a signal enhancer and layer of PDA-AuNPs deposited on GCE as an electrical conductivity enhancer, reproduced from [204]. (C) OTA assay using TiO2 nanotube electrode functionalized with DNA probe tagged with MB, reproduced from [205]. (D) Schematic protocols for photoelectrochemical aptasensor ATZ assay with nuclease signal amplification using TiO2 nanotube covered by a layer of AuNPs, further functionalized with ATZ aptamer/graphene mixture as a sensor probe, reproduced from [206].
5. Conclusions and Future Outlook
The distinctive and impressive advantages of aptamers compared with antibodies permit them to be preferred in molecular diagnostics for a wide range of biomarkers. Aptamers have intrinsic advantages, such as their availability for both chemical modifications and conjugation with different labels, facilitating their ability to be used to construct a sensitive and highly selective platform sensor. In this review, recent advances in the different methods of employing aptasensors including colorimetric, fluorometric, and electrochemical strategies were discussed. The simplicity and small sizes of the aptamers combined with the versatile optical properties and large surface areas of nanomaterials leads to a platform with great potential for highly sensitive and selective biological recognition and signal transduction for various analytes such as metals, small molecules, toxins, proteins, cells, and bacteria with lower LODs and high sensitivity and selectivity.
In this review, each detection method with its own different strategies was emphasized briefly with schematic designs and all the aptamer information, including the detection ranges of the discussed aptasensors, summarized in Table 1. Each aptasensor method has advantages, but the limitations of each method have to be considered before assaying the biomarkers. The optical strategies are considered promising assay methods, as a sensitive response could be achieved; however, some limitations should be considered before assigning a suitable strategy. The colorimetric aptasensor sensors are considered more promising for point of case testing (POCT) owing to naked-eye readout; however, their sensitivity fails to meet the required criteria owing to the higher LODs compared to other methods. Moreover, some limitations were demonstrated in the AuNP–salt-induced aggregation, in which the surface of the AuNP was easily accessible by several targets and aptamers, affecting the reliability of the sensor. This hurdle was overcome by using a strong capping agent [93,94,96]. Despite the fact that the fluorescent aptasensor achieved a high sensitivity for the assay of different targets compared to colorimetric, it requires laboratories and clinical centers with infrastructure for achieving POCT diagnosis, and the limitations of the photobleaching of the fluorescent molecules over time, compromising their stability, is considered an obstacle regarding fluorescent aptasensor development. Generally, electrochemical aptasensors are rapid, easy, and higher in sensitivity compared to optical sensors, making them the best candidates for the on-site rapid assay of biomarkers.
Biomarker detection based on aptamer functionalization still needs to overcome these limitations in order to be available for the multi-detection of metal ions, DNA, and proteins. Furthermore, the development of more specific aptamers is still needed, as is also integration into sensor platforms. Aptasensor platforms need more attention regarding (1) simultaneous multiple marker detection; (2) the long-term stability of biosensor assays; (3) direct assays in real sample matrixes; (4) understanding the nature of the binding competition between an aptamer and target on the surface of a nanomaterial, which could affect the sensor reliability; (5) more focus on the future development of in vivo aptamer sensing technology, the possible problems, and their solutions; and (6) more intensive research regarding the improvement of the POCT of biomarkers using aptasensors. The aforementioned hurdles need to be overcome quickly for the achievement of a reliable and selective detection of markers with ultrasensitivity, with affordable and portable on-site analytical devices. We are sure that the scientific community has the talent to offer solutions to these hurdles in order to design an affordable, easy-to-use nanomaterial-based electro-optical aptasensor integrated with rolling-cycle amplification technology.
Acknowledgments
Samy M. Shaban acknowledges the support from Egyptian Petroleum Research Institute.
Funding
This research was funded by the National Research Foundation of Korea (NRF) (2018R1D1A1B07041584) and the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2020R1A5A1018052).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nat. Cell Biol. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Stoltenburg R., Reinemann C., Strehlitz B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Zhuo Z., Yu Y., Wang M., Li J., Zhang Z.-K., Liu J., Wu X., Lu A., Zhang G., Zhang B.-T. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017;18:2142. doi: 10.3390/ijms18102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y.X., Kwon Y.J. Aptamers: The “evolution” of SELEX. Methods. 2016;106:21–28. doi: 10.1016/j.ymeth.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Ohuchi S. Cell-SELEX Technology. BioRes. Open Access. 2012;1:265–272. doi: 10.1089/biores.2012.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi X., Yan X., Zhao Y., Li L., Wang S. Highly sensitive and specific detection of small molecules using advanced aptasensors based on split aptamers: A review. TrAC Trends Anal. Chem. 2020;133:116069. doi: 10.1016/j.trac.2020.116069. [DOI] [Google Scholar]
- 8.Xu L., Duan W., Chen F., Zhang J., Li H. A photoelectrochemical aptasensor for the determination of bisphenol A based on the Cu (I) modified graphitic carbon nitride. J. Hazard. Mater. 2020;400:123162. doi: 10.1016/j.jhazmat.2020.123162. [DOI] [PubMed] [Google Scholar]
- 9.Ojha Y.R., Giovannucci D.R., Cameron B.D. Selection and characterization of structure-switching DNA aptamers for the salivary peptide histatin 3. J. Biotechnol. 2021;327:9–17. doi: 10.1016/j.jbiotec.2020.12.018. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z., Yuan Y., Wu X., Ning Q., Wu S., Fu L. A turn-off colorimetric DNAzyme-aptasensor for ultra-high sensitive detection of viable Cronobacter sakazakii. Sens. Actuators B Chem. 2020;322:128646. doi: 10.1016/j.snb.2020.128646. [DOI] [Google Scholar]
- 11.Chen H., Park S.-G., Choi N., Moon J.-I., Dang H., Das A., Lee S., Kim D.-G., Chen L., Choo J. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020;167:112496. doi: 10.1016/j.bios.2020.112496. [DOI] [PubMed] [Google Scholar]
- 12.Fan Y.-Y., Deng X., Wang M., Li J., Zhang Z.-Q. A dual-function oligonucleotide-based ratiometric fluorescence sensor for ATP detection. Talanta. 2020;219:121349. doi: 10.1016/j.talanta.2020.121349. [DOI] [PubMed] [Google Scholar]
- 13.Ren Q., Mou J., Guo Y., Wang H., Cao X., Zhang F., Xia J., Wang Z. Simple homogeneous electrochemical target-responsive aptasensor based on aptamer bio-gated and porous carbon nanocontainer derived from ZIF-8. Biosens. Bioelectron. 2020;166:112448. doi: 10.1016/j.bios.2020.112448. [DOI] [PubMed] [Google Scholar]
- 14.Dong H., Chen H., Jiang J., Zhang H., Cai C., Shen Q. Highly Sensitive Electrochemical Detection of Tumor Exosomes Based on Aptamer Recognition-Induced Multi-DNA Release and Cyclic Enzymatic Amplification. Anal. Chem. 2018;90:4507–4513. doi: 10.1021/acs.analchem.7b04863. [DOI] [PubMed] [Google Scholar]
- 15.Jepsen M.D., Sparvath S.M., Nielsen T.B., Langvad A.H., Grossi G., Gothelf K.V., Andersen E.S. Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat. Commun. 2018;9:18. doi: 10.1038/s41467-017-02435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kun Q., Lin Y., Peng H., Cheng L., Cui H., Hong N., Xiong J., Fan H. A “signal-on” switch electrochemiluminescence biosensor for the detection of tumor cells. J. Electroanal. Chem. 2018;808:101–106. doi: 10.1016/j.jelechem.2017.11.031. [DOI] [Google Scholar]
- 17.Huang K., Doyle F., Wurz Z.E., Tenenbaum S.A., Hammond R.K., Caplan J., Meyers B.C. FASTmiR: An RNA-based sensor for in vitro quantification and live-cell localization of small RNAs. Nucleic Acids Res. 2017;45:e130. doi: 10.1093/nar/gkx504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Cheng F.-F., Zheng T., Zhu J.-J. Versatile aptasensor for electrochemical quantification of cell surface glycan and naked-eye tracking glycolytic inhibition in living cells. Biosens. Bioelectron. 2017;89:937–945. doi: 10.1016/j.bios.2016.09.087. [DOI] [PubMed] [Google Scholar]
- 19.Mao Y., Chen Y., Li S., Lin S., Jiang Y. A Graphene-Based Biosensing Platform Based on Regulated Release of an Aptameric DNA Biosensor. Sensors. 2015;15:28244–28256. doi: 10.3390/s151128244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fellows T., Ho L., Flanagan S., Fogel R., Ojo D., Limson J. Gold nanoparticle-streptavidin conjugates for rapid and efficient screening of aptamer function in lateral flow sensors using novel CD4-binding aptamers identified through Crossover-SELEX. Analyst. 2020;145:5180–5193. doi: 10.1039/D0AN00634C. [DOI] [PubMed] [Google Scholar]
- 21.Shafiei F., McAuliffe K., Bagheri Y., Sun Z., Yu Q., Wu R., You M. Paper-based fluorogenic RNA aptamer sensors for label-free detection of small molecules. Anal. Methods. 2020;12:2674–2681. doi: 10.1039/D0AY00588F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisi F., Peterson J.R., Gooding J.J. The application of personal glucose meters as universal point-of-care diagnostic tools. Biosens. Bioelectron. 2020;148:111835. doi: 10.1016/j.bios.2019.111835. [DOI] [PubMed] [Google Scholar]
- 23.Li Z.-H., Mohamed M.A., Mohan A., Zhu Z., Sharma V., Mishra G.K., Mishra R.K. Application of Electrochemical Aptasensors toward Clinical Diagnostics, Food, and Environmental Monitoring: Review. Sensors. 2019;19:5435. doi: 10.3390/s19245435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaban S.M., Abd-Elaal A.A. Studying the silver nanoparticles influence on thermodynamic behavior and antimicrobial activities of novel amide Gemini cationic surfactants. Mater. Sci. Eng. C. 2017;76:871–885. doi: 10.1016/j.msec.2017.03.185. [DOI] [PubMed] [Google Scholar]
- 25.Shaban S.M., Lee J.-Y., Kim D.-H. Dual-Surfactant-Capped Ag Nanoparticles as a Highly Selective and Sensitive Colorimetric Sensor for Citrate Detection. ACS Omega. 2020;5:10696–10703. doi: 10.1021/acsomega.9b04199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaban S.M., Kim D.H. The influence of the Gemini surfactants hydrocarbon tail on in-situ synthesis of silver nanoparticles: Characterization, surface studies and biological performance. Korean J. Chem. Eng. 2020;37:1008–1019. doi: 10.1007/s11814-020-0542-1. [DOI] [Google Scholar]
- 27.Hussain F., Shaban S.M., Kim J., Kim Y.-J. One-pot synthesis of highly stable and concentrated silver nanoparticles with enhanced catalytic activity. Korean J. Chem. Eng. 2019;36:988–995. doi: 10.1007/s11814-019-0270-6. [DOI] [Google Scholar]
- 28.Badr E.A., Hefni H.H., Shafek S., Shaban S.M. Synthesis of anionic chitosan surfactant and application in silver nanoparticles preparation and corrosion inhibition of steel. Int. J. Biol. Macromol. 2020;157:187–201. doi: 10.1016/j.ijbiomac.2020.04.184. [DOI] [PubMed] [Google Scholar]
- 29.Aiad I., Shaban S.M., Tawfik S.M., Khalil M.M., El-Wakeel N. Effect of some prepared surfactants on silver nanoparticles formation and surface solution behavior and their biological activity. J. Mol. Liq. 2018;266:381–392. doi: 10.1016/j.molliq.2018.06.089. [DOI] [Google Scholar]
- 30.Shaban S.M., Aiad I., Yassin F.A., Mosalam A. The Tail Effect of Some Prepared Cationic Surfactants on Silver Nanoparticle Preparation and Their Surface, Thermodynamic Parameters, and Antimicrobial Activity. J. Surfactants Deterg. 2019;22:1445–1460. doi: 10.1002/jsde.12318. [DOI] [Google Scholar]
- 31.Sharifi S., Vahed S.Z., Ahmadian E., Dizaj S.M., Eftekhari A., Khalilov R., Ahmadi M., Hamidi-Asl E., Labib M. Detection of pathogenic bacteria via nanomaterials-modified aptasensors. Biosens. Bioelectron. 2020;150:111933. doi: 10.1016/j.bios.2019.111933. [DOI] [PubMed] [Google Scholar]
- 32.Meng T., Jia H., Ye H., Zeng T., Yang X., Wang H., Zhang Y. Facile preparation of CoMoO4 nanorods at macroporous carbon hybrid electrocatalyst for non-enzymatic glucose detection. J. Colloid Interface Sci. 2020;560:1–10. doi: 10.1016/j.jcis.2019.10.054. [DOI] [PubMed] [Google Scholar]
- 33.Li B., Luo J., Huang X., Lin L., Wang L., Hu M., Tang L., Xue H., Gao J., Mai Y.-W. A highly stretchable, super-hydrophobic strain sensor based on polydopamine and graphene reinforced nanofiber composite for human motion monitoring. Compos. Part B Eng. 2020;181:107580. doi: 10.1016/j.compositesb.2019.107580. [DOI] [Google Scholar]
- 34.Iranmanesh T., Foroughi M.M., Jahani S., Zandi M.S., Nadiki H.H. Green and facile microwave solvent-free synthesis of CeO2 nanoparticle-decorated CNTs as a quadruplet electrochemical platform for ultrasensitive and simultaneous detection of ascorbic acid, dopamine, uric acid and acetaminophen. Talanta. 2020;207:120318. doi: 10.1016/j.talanta.2019.120318. [DOI] [PubMed] [Google Scholar]
- 35.Shetti N.P., Bukkitgar S.D., Reddy K.R., Reddy C.V., Aminabhavi T.M. ZnO-based nanostructured electrodes for electrochemical sensors and biosensors in biomedical applications. Biosens. Bioelectron. 2019;141:111417. doi: 10.1016/j.bios.2019.111417. [DOI] [PubMed] [Google Scholar]
- 36.Deshmukh S., Patil S., Mullani S., Delekar S.D. Silver nanoparticles as an effective disinfectant: A review. Mater. Sci. Eng. C. 2019;97:954–965. doi: 10.1016/j.msec.2018.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han S., Yang L., Wen Z., Chu S., Wang M., Wang Z., Jiang C. A dual-response ratiometric fluorescent sensor by europium-doped CdTe quantum dots for visual and colorimetric detection of tetracycline. J. Hazard. Mater. 2020;398:122894. doi: 10.1016/j.jhazmat.2020.122894. [DOI] [PubMed] [Google Scholar]
- 38.Kong D., Yao J., Li X., Luo J., Yang M. A reusable AuNPS with increased stability applied for fast screening of trace heavy metals in edible and medicinal marine products. Ecotoxicol. Environ. Saf. 2020;204:111107. doi: 10.1016/j.ecoenv.2020.111107. [DOI] [PubMed] [Google Scholar]
- 39.Cheng N., Song Y., Fu Q., Du D., Luo Y., Wang Y., Xu W., Lin Y. Aptasensor based on fluorophore-quencher nano-pair and smartphone spectrum reader for on-site quantification of multi-pesticides. Biosens. Bioelectron. 2018;117:75–83. doi: 10.1016/j.bios.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Sha H., Ke H., Xiong X., Jia N. A sandwich-type electrochemiluminescence aptasensor for insulin detection based on the nano-C60/BSA@luminol nanocomposite and ferrocene derivative. Electrochim. Acta. 2018;290:90–97. doi: 10.1016/j.electacta.2018.08.080. [DOI] [Google Scholar]
- 41.Wei B., Mao K., Liu N., Zhang M., Yang Z. Graphene nanocomposites modified electrochemical aptamer sensor for rapid and highly sensitive detection of prostate specific antigen. Biosens. Bioelectron. 2018;121:41–46. doi: 10.1016/j.bios.2018.08.067. [DOI] [PubMed] [Google Scholar]
- 42.Arvand M., Mirroshandel A.A. An efficient fluorescence resonance energy transfer system from quantum dots to graphene oxide nano sheets: Application in a photoluminescence aptasensing probe for the sensitive detection of diazinon. Food Chem. 2019;280:115–122. doi: 10.1016/j.foodchem.2018.12.069. [DOI] [PubMed] [Google Scholar]
- 43.He B., Dong X. Hierarchically porous Zr-MOFs labelled methylene blue as signal tags for electrochemical patulin aptasensor based on ZnO nano flower. Sens. Actuators B Chem. 2019;294:192–198. doi: 10.1016/j.snb.2019.05.045. [DOI] [Google Scholar]
- 44.Sarabaegi M., Roushani M. A nano-sized chitosan particle based electrochemical aptasensor for sensitive detection of P. aeruginosa. Anal. Methods. 2019;11:5591–5597. doi: 10.1039/C9AY01509D. [DOI] [Google Scholar]
- 45.Wang J., Guo X., Liu R., Guo J., Zhang Y., Zhang W., Sang S. Detection of carcinoembryonic antigen using a magnetoelastic nano-biosensor amplified with DNA-templated silver nanoclusters. Nanotechnology. 2019;31:015501. doi: 10.1088/1361-6528/ab4506. [DOI] [PubMed] [Google Scholar]
- 46.Abazar F., Noorbakhsh A. Chitosan-carbon quantum dots as a new platform for highly sensitive insulin impedimetric aptasensor. Sens. Actuators B Chem. 2020;304:127281. doi: 10.1016/j.snb.2019.127281. [DOI] [Google Scholar]
- 47.He Z.-J., Kang T.-F., Lu L.-P., Cheng S.-Y. An electrochemiluminescence aptamer sensor for chloramphenicol based on GO-QDs nanocomposites and enzyme-linked aptamers. J. Electroanal. Chem. 2020;860:113870. doi: 10.1016/j.jelechem.2020.113870. [DOI] [Google Scholar]
- 48.Walter J.-G., Eilers A., Alwis L.S.M., Roth B., Bremer K. SPR Biosensor Based on Polymer Multi-Mode Optical Waveguide and Nanoparticle Signal Enhancement. Sensors. 2020;20:2889. doi: 10.3390/s20102889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pla L., Santiago-Felipe S., Tormo-Mas M., Pemán J., Sancenón F., Aznar E., Martínez-Máñez R. Aptamer-Capped nanoporous anodic alumina for Staphylococcus aureus detection. Sens. Actuators B Chem. 2020;320:128281. doi: 10.1016/j.snb.2020.128281. [DOI] [Google Scholar]
- 50.Wang Z., Lu Y. Functional DNA directed assembly of nanomaterials for biosensing. J. Mater. Chem. 2009;19:1788–1798. doi: 10.1039/b813939c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu Y., Liu J. Functional DNA nanotechnology: Emerging applications of DNAzymes and aptamers. Curr. Opin. Biotechnol. 2006;17:580–588. doi: 10.1016/j.copbio.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Farzin L., Shamsipur M., Sheibani S. A review: Aptamer-based analytical strategies using the nanomaterials for environmental and human monitoring of toxic heavy metals. Talanta. 2017;174:619–627. doi: 10.1016/j.talanta.2017.06.066. [DOI] [PubMed] [Google Scholar]
- 53.Zhu C., Yang G., Li H., Du D., Lin Y. Electrochemical Sensors and Biosensors Based on Nanomaterials and Nanostructures. Anal. Chem. 2014;87:230–249. doi: 10.1021/ac5039863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goyal R.N., Gupta V.K., Chatterjee S. Voltammetric biosensors for the determination of paracetamol at carbon nanotube modified pyrolytic graphite electrode. Sens. Actuators B Chem. 2010;149:252–258. doi: 10.1016/j.snb.2010.05.019. [DOI] [Google Scholar]
- 55.Urmann K., Modrejewski J., Scheper P.T., Walter J.-G. Aptamer-modified nanomaterials: Principles and applications. BioNanoMaterials. 2016;18 doi: 10.1515/bnm-2016-0012. [DOI] [Google Scholar]
- 56.Xiao Z., Farokhzad O.C. Aptamer-Functionalized Nanoparticles for Medical Applications: Challenges and Opportunities. ACS Nano. 2012;6:3670–3676. doi: 10.1021/nn301869z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yue F., Li F., Kong Q., Guo Y., Sun X. Recent advances in aptamer-based sensors for aminoglycoside antibiotics detection and their applications. Sci. Total Environ. 2021;762:143129. doi: 10.1016/j.scitotenv.2020.143129. [DOI] [PubMed] [Google Scholar]
- 58.Zhang N., Liu B., Cui X., Li Y., Tang J., Wang H., Zhang D., Li Z. Recent Advances in Aptasensors for Mycotoxin Detection: On the Surface and in the Colloid. Talanta. 2021;223:121729. doi: 10.1016/j.talanta.2020.121729. [DOI] [PubMed] [Google Scholar]
- 59.Ghorbani F., Abbaszadeh H., Dolatabadi J.E.N., Aghebati-Maleki L., Yousefi M. Application of various optical and electrochemical aptasensors for detection of human prostate specific antigen: A review. Biosens. Bioelectron. 2019;142:111484. doi: 10.1016/j.bios.2019.111484. [DOI] [PubMed] [Google Scholar]
- 60.Rajabnejad S.-H., Badibostan H., Verdian A., Karimi G.R., Fooladi E., Feizy J. Aptasensors as promising new tools in bisphenol A detection—An invisible pollution in food and environment. Microchem. J. 2020;155:104722. doi: 10.1016/j.microc.2020.104722. [DOI] [Google Scholar]
- 61.Zhuang Y., Liu L., Wu X., Tian Y., Zhou X., Xu S., Xie Z., Ma Y. Size and Shape Effect of Gold Nanoparticles in “Far-Field” Surface Plasmon Resonance. Part. Part. Syst. Charact. 2019;36:1800077. doi: 10.1002/ppsc.201800077. [DOI] [Google Scholar]
- 62.El-Brolossy T.A., Abdallah T., Mohamed M.B., Easawi K., Negm S., Talaat H. Shape and size dependence of the surface plasmon resonance of gold nanoparticles studied by Photoacoustic technique. Eur. Phys. J. Spéc. Top. 2008;153:361–364. doi: 10.1140/epjst/e2008-00462-0. [DOI] [Google Scholar]
- 63.Bin Jeon H., Tsalu P.V., Ha J.W. Shape Effect on the Refractive Index Sensitivity at Localized Surface Plasmon Resonance Inflection Points of Single Gold Nanocubes with Vertices. Sci. Rep. 2019;9:13635. doi: 10.1038/s41598-019-50032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang D., Zakaria S., Deng M., Allen N., Tram K., Li Y. Integrating Deoxyribozymes into Colorimetric Sensing Platforms. Sensors. 2016;16:2061. doi: 10.3390/s16122061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shalkevich N., Escher W., Bürgi T., Michel B., Si-Ahmed L., Poulikakos D. On the Thermal Conductivity of Gold Nanoparticle Colloids. Langmuir. 2010;26:663–670. doi: 10.1021/la9022757. [DOI] [PubMed] [Google Scholar]
- 66.Jeon H., Lee K. Effect of gold nanoparticle morphology on thermal properties of polyimide nanocomposite films. Coll. Surf. A Physicochem. Eng. Asp. 2019;579:123651. doi: 10.1016/j.colsurfa.2019.123651. [DOI] [Google Scholar]
- 67.Sunil J., Alex S., Pravin A.A., Pooja M.D., Ginil R. Thermal properties of aqueous silver nanoparticle dispersion. Mater. Today Proc. 2020 doi: 10.1016/j.matpr.2020.04.069. [DOI] [Google Scholar]
- 68.Lucas T.M., Moiseeva E.V., Zhang G., Gobin A.M., Harnett C.K. Thermal properties of infrared absorbent gold nanoparticle coatings for MEMS applications. Sens. Actuators A Phys. 2013;198:81–86. doi: 10.1016/j.sna.2013.04.033. [DOI] [Google Scholar]
- 69.Taghdisi S.M., Danesh N.M., Lavaee P., Ramezani M., Abnous K. An aptasensor for selective, sensitive and fast detection of lead (II) based on polyethyleneimine and gold nanoparticles. Environ. Toxicol. Pharmacol. 2015;39:1206–1211. doi: 10.1016/j.etap.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 70.Priyadarshni N., Nath P., Hanumaiah N., Chanda N. DMSA-Functionalized Gold Nanorod on Paper for Colorimetric Detection and Estimation of Arsenic (III and V) Contamination in Groundwater. ACS Sustain. Chem. Eng. 2018;6:6264–6272. doi: 10.1021/acssuschemeng.8b00068. [DOI] [Google Scholar]
- 71.Li H., Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. USA. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo X.-L., Yuan D.-D., Song T., Li X. DNA nanopore functionalized with aptamer and cell-penetrating peptide for tumor cell recognition. Anal. Bioanal. Chem. 2017;4:3789–3797. doi: 10.1007/s00216-017-0321-y. [DOI] [PubMed] [Google Scholar]
- 73.Guo L., Jackman J.A., Yang H.-H., Chen P., Cho N.-J., Kim D.-H. Strategies for enhancing the sensitivity of plasmonic nanosensors. Nano Today. 2015;10:213–239. doi: 10.1016/j.nantod.2015.02.007. [DOI] [Google Scholar]
- 74.Puiu M., Bala C. SPR and SPR Imaging: Recent Trends in Developing Nanodevices for Detection and Real-Time Monitoring of Biomolecular Events. Sensors. 2016;16:870. doi: 10.3390/s16060870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nusz G.J., Curry A.C., Marinakos S.M., Wax A., Chilkoti A. Rational Selection of Gold Nanorod Geometry for Label-Free Plasmonic Biosensors. ACS Nano. 2009;3:795–806. doi: 10.1021/nn8006465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zijlstra P., Paulo P.M.R., Orrit M. Optical detection of single non-absorbing molecules using the surface plasmon resonance of a gold nanorod. Nat. Nanotechnol. 2012;7:379–382. doi: 10.1038/nnano.2012.51. [DOI] [PubMed] [Google Scholar]
- 77.Xu W., Xue X., Li T., Zeng H., Liu X. Ultrasensitive and Selective Colorimetric DNA Detection by Nicking Endonuclease Assisted Nanoparticle Amplification. Angew. Chem. Int. Ed. 2009;48:6849–6852. doi: 10.1002/anie.200901772. [DOI] [PubMed] [Google Scholar]
- 78.Li J., Deng T., Chu X., Tan W., Jiang J.-H., Shen G., Yu R. Rolling Circle Amplification Combined with Gold Nanoparticle Aggregates for Highly Sensitive Identification of Single-Nucleotide Polymorphisms. Anal. Chem. 2010;82:2811–2816. doi: 10.1021/ac100336n. [DOI] [PubMed] [Google Scholar]
- 79.Xu W., Xie X., Li D., Yang Z., Li T., Liu X. Ultrasensitive Colorimetric DNA Detection using a Combination of Rolling Circle Amplification and Nicking Endonuclease-Assisted Nanoparticle Amplification (NEANA) Small. 2012;8:1846–1850. doi: 10.1002/smll.201200263. [DOI] [PubMed] [Google Scholar]
- 80.Zhou W., Huang P.-J.J., Ding J., Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139:2627–2640. doi: 10.1039/c4an00132j. [DOI] [PubMed] [Google Scholar]
- 81.Niazov T., Pavlov V., Xiao Y., Gill A.R., Willner I. DNAzyme-Functionalized Au Nanoparticles for the Amplified Detection of DNA or Telomerase Activity. Nano Lett. 2004;4:1683–1687. doi: 10.1021/nl0491428. [DOI] [Google Scholar]
- 82.Weizmann Y., Beissenhirtz M.K., Cheglakov Z., Nowarski R., Kotler M., Willner I. A Virus Spotlighted by an Autonomous DNA Machine. Angew. Chem. Int. Ed. 2006;45:7384–7388. doi: 10.1002/anie.200602754. [DOI] [PubMed] [Google Scholar]
- 83.Yang C., Lates V., Prieto-Simón B., Marty J.L., Yang X. Aptamer-DNAzyme hairpins for biosensing of Ochratoxin A. Biosens. Bioelectron. 2012;32:208–212. doi: 10.1016/j.bios.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 84.Gupta R., Kumar A., Kumar S., Pinnaka A.K., Singhal N.K. Naked eye colorimetric detection of Escherichia coli using aptamer conjugated graphene oxide enclosed Gold nanoparticles. Sens. Actuators B Chem. 2021;329:129100. doi: 10.1016/j.snb.2020.129100. [DOI] [Google Scholar]
- 85.Lai X., Zhang S., Du G., Wang Y., Han Y., Ye N., Xiang Y. Ultrasensitive Determination of Malathion in Apples by Aptamer-Based Resonance Scattering. Anal. Lett. 2020:1–15. doi: 10.1080/00032719.2020.1820022. [DOI] [Google Scholar]
- 86.Xu L., Liang J., Wang Y., Ren S., Wu J., Zhou H., Gao Z. Highly Selective, Aptamer-Based, Ultrasensitive Nanogold Colorimetric Smartphone Readout for Detection of Cd (II) Molecules. 2019;24:2745. doi: 10.3390/molecules24152745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen S., Yang X., Fu S., Qin X., Yang T., Man C., Jiang Y. A novel AuNPs colorimetric sensor for sensitively detecting viable Salmonella typhimurium based on dual aptamers. Food Control. 2020;115:107281. doi: 10.1016/j.foodcont.2020.107281. [DOI] [Google Scholar]
- 88.Yi J., Wu P., Li G., Xiao W., Li L., He Y., He Y., Ding P., Chen C. A composite prepared from carboxymethyl chitosan and aptamer-modified gold nanoparticles for the colorimetric determination of Salmonella typhimurium. Microchim. Acta. 2019;186:711. doi: 10.1007/s00604-019-3827-5. [DOI] [PubMed] [Google Scholar]
- 89.Jalalian S.H., Lavaee P., Ramezani M., Danesh N.M., Alibolandi M., Abnous K., Taghdisi S.M. An optical aptasensor for aflatoxin M1 detection based on target-induced protection of gold nanoparticles against salt-induced aggregation and silica nanoparticles. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021;246:119062. doi: 10.1016/j.saa.2020.119062. [DOI] [PubMed] [Google Scholar]
- 90.Lerdsri J., Chananchana W., Upan J., Sridara T., Jakmunee J. Label-free colorimetric aptasensor for rapid detection of aflatoxin B1 by utilizing cationic perylene probe and localized surface plasmon resonance of gold nanoparticles. Sens. Actuators B Chem. 2020;320:128356. doi: 10.1016/j.snb.2020.128356. [DOI] [Google Scholar]
- 91.Liu X., He F., Zhang F., Zhang Z., Huang Z., Liu J. Dopamine and Melamine Binding to Gold Nanoparticles Dominates Their Aptamer-Based Label-Free Colorimetric Sensing. Anal. Chem. 2020;92:9370–9378. doi: 10.1021/acs.analchem.0c01773. [DOI] [PubMed] [Google Scholar]
- 92.Zong C., Liu J. The Arsenic-Binding Aptamer Cannot Bind Arsenic: Critical Evaluation of Aptamer Selection and Binding. Anal. Chem. 2019;91:10887–10893. doi: 10.1021/acs.analchem.9b02789. [DOI] [PubMed] [Google Scholar]
- 93.Cao R., Li B., Zhang Y., Zhang Z. Naked-eye sensitive detection of nuclease activity using positively-charged gold nanoparticles as colorimetric probes. Chem. Commun. 2011;47:12301–12303. doi: 10.1039/c1cc15994a. [DOI] [PubMed] [Google Scholar]
- 94.Bian X., Song Z.-L., Qian Y., Gao W., Cheng Z.-Q., Chen L., Liang H., Ding D., Nie X.-K., Chen Z., et al. Fabrication of Graphene-isolated-Au-nanocrystal Nanostructures for Multimodal Cell Imaging and Photothermal-enhanced Chemotherapy. Sci. Rep. 2014;4:6093. doi: 10.1038/srep06093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qi Y., Chen Y., Xiu F.-R., Hou J. An aptamer-based colorimetric sensing of acetamiprid in environmental samples: Convenience, sensitivity and practicability. Sens. Actuators B Chem. 2020;304:127359. doi: 10.1016/j.snb.2019.127359. [DOI] [Google Scholar]
- 96.Qi Y., Ma J., Chen X., Xiu F.-R., Chen Y., Lu Y. Practical aptamer-based assay of heavy metal mercury ion in contaminated environmental samples: Convenience and sensitivity. Anal. Bioanal. Chem. 2019;412:439–448. doi: 10.1007/s00216-019-02253-8. [DOI] [PubMed] [Google Scholar]
- 97.Chen Q., Gao R., Jia L. Enhancement of the peroxidase-like activity of aptamers modified gold nanoclusters by bacteria for colorimetric detection of Salmonella typhimurium. Talanta. 2021;221:121476. doi: 10.1016/j.talanta.2020.121476. [DOI] [PubMed] [Google Scholar]
- 98.Wang S., Zhang H., Li W., Birech Z., Ma L., Li D., Li S., Wang L., Shang J., Hu J. A multi-channel localized surface plasmon resonance system for absorptiometric determination of abscisic acid by using gold nanoparticles functionalized with a polyadenine-tailed aptamer. Microchim. Acta. 2019;187:20. doi: 10.1007/s00604-019-4003-7. [DOI] [PubMed] [Google Scholar]
- 99.Jo S., Lee W., Park J., Kim W., Kim W., Lee G., Hong J., Park J. Localized surface plasmon resonance aptasensor for the highly sensitive direct detection of cortisol in human saliva. Sens. Actuators B Chem. 2020;304:127424. doi: 10.1016/j.snb.2019.127424. [DOI] [Google Scholar]
- 100.Lee W., Shaban S.M., Pyun D.G., Kim Y.-J. Solid-phase colorimetric apta-biosensor for thrombin detection. Thin Solid Films. 2019;686:137428. doi: 10.1016/j.tsf.2019.137428. [DOI] [Google Scholar]
- 101.Tao Z., Wei L., Wu S., Duan N., Li X., Wang Z. A colorimetric aptamer-based method for detection of cadmium using the enhanced peroxidase-like activity of Au–MoS2 nanocomposites. Anal. Biochem. 2020;608:113844. doi: 10.1016/j.ab.2020.113844. [DOI] [PubMed] [Google Scholar]
- 102.Yan X.-L., Xue X.-X., Luo J., Jian Y.-T., Tong L., Zheng X.-J. Construction of chemiluminescence aptasensor platform using magnetic microsphere for ochratoxin A detection based on G bases derivative reaction and Au NPs catalyzing luminol system. Sens. Actuators B Chem. 2020;320:128375. doi: 10.1016/j.snb.2020.128375. [DOI] [Google Scholar]
- 103.Miao X., Zhu Z., Jia H., Lu C., Liu X., Mao D., Chen G. Colorimetric detection of cancer biomarker based on enzyme enrichment and pH sensing. Sens. Actuators B Chem. 2020;320:128435. doi: 10.1016/j.snb.2020.128435. [DOI] [Google Scholar]
- 104.Zhang G., Zhang L., Yu Y., Lin B., Wang Y., Guo M., Cao Y. Dual-mode of electrochemical-colorimetric imprinted sensing strategy based on self-sacrifice beacon for diversified determination of cardiac troponin I in serum. Biosens. Bioelectron. 2020;167:112502. doi: 10.1016/j.bios.2020.112502. [DOI] [PubMed] [Google Scholar]
- 105.Wang J., Wang Q., Zhong Y., Wu D., Gan N. A sandwich-type aptasensor for point-of-care measurements of low-density lipoprotein in plasma based on aptamer-modified MOF and magnetic silica composite probes. Microchem. J. 2020;158:105288. doi: 10.1016/j.microc.2020.105288. [DOI] [Google Scholar]
- 106.Li S., Zhao X., Yu X., Wan Y., Yin M., Zhang W., Cao B., Wang H. Fe3O4 Nanozymes with Aptamer-Tuned Catalysis for Selective Colorimetric Analysis of ATP in Blood. Anal. Chem. 2019;91:14737–14742. doi: 10.1021/acs.analchem.9b04116. [DOI] [PubMed] [Google Scholar]
- 107.Tao Z., Zhou Y., Duan N., Wang Z. A Colorimetric Aptamer Sensor Based on the Enhanced Peroxidase Activity of Functionalized Graphene/Fe3O4-AuNPs for Detection of Lead (II) Ions. Catalysts. 2020;10:600. doi: 10.3390/catal10060600. [DOI] [Google Scholar]
- 108.Duan N., Yang W., Wu S., Zou Y., Wang Z. A Visual and Sensitive Detection of Escherichia coli Based on Aptamer and Peroxidase-like Mimics of Copper-Metal Organic Framework Nanoparticles. Food Anal. Methods. 2020;13:1433–1441. doi: 10.1007/s12161-020-01765-9. [DOI] [Google Scholar]
- 109.Huang Y., Zhang L., Li Z., Gopinath S.C., Chen Y., Xiao Y. Aptamer–17β-estradiol–antibody sandwich ELISA for determination of gynecological endocrine function. Biotechnol. Appl. Biochem. 2020 doi: 10.1002/bab.2008. [DOI] [PubMed] [Google Scholar]
- 110.Xing K.-Y., Peng J., Shan S., Liu D.-F., Huang Y.-N., Lai W. Green Enzyme-Linked Immunosorbent Assay Based on the Single-Stranded Binding Protein-Assisted Aptamer for the Detection of Mycotoxin. Anal. Chem. 2020;92:8422–8426. doi: 10.1021/acs.analchem.0c01073. [DOI] [PubMed] [Google Scholar]
- 111.Wei Y., Wang D., Zhang Y., Sui J., Xu Z.-R. Multicolor and photothermal dual-readout biosensor for visual detection of prostate specific antigen. Biosens. Bioelectron. 2019;140:111345. doi: 10.1016/j.bios.2019.111345. [DOI] [PubMed] [Google Scholar]
- 112.Li T., Ou G., Chen X., Li Z., Hu R., Li Y., Yang Y., Liu M. Naked-eye based point-of-care detection of E.coli O157: H7 by a signal-amplified microfluidic aptasensor. Anal. Chim. Acta. 2020;1130:20–28. doi: 10.1016/j.aca.2020.07.031. [DOI] [PubMed] [Google Scholar]
- 113.Pehlivan Z.S., Torabfam M., Kurt H., Ow-Yang C., Hildebrandt N., Yüce M. Aptamer and nanomaterial based FRET biosensors: A review on recent advances (2014–2019) Microchim. Acta. 2019;186:563. doi: 10.1007/s00604-019-3659-3. [DOI] [PubMed] [Google Scholar]
- 114.Şahin S., Caglayan M.O., Üstundağ Z. A review on nanostructure-based mercury (II) detection and monitoring focusing on aptamer and oligonucleotide biosensors. Talanta. 2020;220:121437. doi: 10.1016/j.talanta.2020.121437. [DOI] [PubMed] [Google Scholar]
- 115.Li F., Pei H., Wang L., Lu J., Gao J., Jiang B., Zhao X., Fan C. Nanomaterial-Based Fluorescent DNA Analysis: A Comparative Study of the Quenching Effects of Graphene Oxide, Carbon Nanotubes, and Gold Nanoparticles. Adv. Funct. Mater. 2013;23:4140–4148. doi: 10.1002/adfm.201203816. [DOI] [Google Scholar]
- 116.Chen L., Lee S., Lee M., Lim C., Choo J., Park J.Y., Lee S., Joo S.-W., Lee K.-H., Choi Y.-W. DNA hybridization detection in a microfluidic channel using two fluorescently labelled nucleic acid probes. Biosens. Bioelectron. 2008;23:1878–1882. doi: 10.1016/j.bios.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 117.Shi J., Tian F., Lyu J., Yang M. Nanoparticle based fluorescence resonance energy transfer (FRET) for biosensing applications. J. Mater. Chem. B. 2015;3:6989–7005. doi: 10.1039/C5TB00885A. [DOI] [PubMed] [Google Scholar]
- 118.Wang L., Zhu F., Chen M., Zhu Y., Xiao J., Yang H., Chen X. Rapid and visual detection of aflatoxin B1 in foodstuffs using aptamer/G-quadruplex DNAzyme probe with low background noise. Food Chem. 2019;271:581–587. doi: 10.1016/j.foodchem.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 119.Khan I.M., Niazi S., Yu Y., Pasha I., Yue L., Mohsin A., Shoaib M., Iqbal M.W., Khaliq A., Wang Z. Fabrication of PAA coated green-emitting AuNCs for construction of label-free FRET assembly for specific recognition of T-2 toxin. Sens. Actuators B Chem. 2020;321:128470. doi: 10.1016/j.snb.2020.128470. [DOI] [Google Scholar]
- 120.Shirani M., Kalantari H., Khodayar M.J., Kouchak M., Rahbar N. A novel strategy for detection of small molecules based on aptamer/gold nanoparticles/graphitic carbon nitride nanosheets as fluorescent biosensor. Talanta. 2020;219:121235. doi: 10.1016/j.talanta.2020.121235. [DOI] [PubMed] [Google Scholar]
- 121.Tan X., Wang X., Hao A., Liu Y., Wang X., Chu T., Jiang M., Yang Y., Ming D. Aptamer-based ratiometric fluorescent nanoprobe for specific and visual detection of zearalenone. Microchem. J. 2020;157:104943. doi: 10.1016/j.microc.2020.104943. [DOI] [Google Scholar]
- 122.Guo Y., Zhang J., Zhang W., Hu D. Green fluorescent carbon quantum dots functionalized with polyethyleneimine, and their application to aptamer-based determination of thrombin and ATP. Microchim. Acta. 2019;186:717. doi: 10.1007/s00604-019-3874-y. [DOI] [PubMed] [Google Scholar]
- 123.Jia Y., Zhou G., Wang X., Zhang Y., Li Z., Liu P., Yu B., Zhang J. A metal-organic framework/aptamer system as a fluorescent biosensor for determination of aflatoxin B1 in food samples. Talanta. 2020;219:121342. doi: 10.1016/j.talanta.2020.121342. [DOI] [PubMed] [Google Scholar]
- 124.Li D., Guo J., Zhao L., Zhang G., Yan G. A label-free RTP sensor based on aptamer/quantum dot nanocomposites for cytochrome c detection. RSC Adv. 2019;9:31953–31959. doi: 10.1039/C9RA05761G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xiong J., Li S., Li Y., Chen Y., Liu Y., Gan J., Ju J., Xian Y., Xiong X. Fluorescent Aptamer-Polyethylene Glycol Functionalized Graphene Oxide Biosensor for Profenofos Detection in Food. Chem. Res. Chin. Univ. 2019;36:787–794. doi: 10.1007/s40242-019-9257-4. [DOI] [Google Scholar]
- 126.Pang S., Liu S., Su X. An ultrasensitive sensing strategy for the detection of lead (II) ions based on the intermolecular G-quadruplex and graphene oxide. Sens. Actuators B Chem. 2015;208:415–420. doi: 10.1016/j.snb.2014.11.030. [DOI] [Google Scholar]
- 127.Qian Z.S., Shan X.Y., Chai L.J., Chen J.R., Feng H. A fluorescent nanosensor based on graphene quantum dots–aptamer probe and graphene oxide platform for detection of lead (II) ion. Biosens. Bioelectron. 2015;68:225–231. doi: 10.1016/j.bios.2014.12.057. [DOI] [PubMed] [Google Scholar]
- 128.Li M., Zhou X., Guo S., Wu N. Detection of lead (II) with a “turn-on” fluorescent biosensor based on energy transfer from CdSe/ZnS quantum dots to graphene oxide. Biosens. Bioelectron. 2013;43:69–74. doi: 10.1016/j.bios.2012.11.039. [DOI] [PubMed] [Google Scholar]
- 129.Jia Y., Wu F., Liu P., Zhou G., Yu B., Lou X., Xia F. A label-free fluorescent aptasensor for the detection of Aflatoxin B1 in food samples using AIEgens and graphene oxide. Talanta. 2019;198:71–77. doi: 10.1016/j.talanta.2019.01.078. [DOI] [PubMed] [Google Scholar]
- 130.Khan R., Sherazi T.A., Catanante G., Rasheed S., Marty J.L., Hayat A. Switchable fluorescence sensor toward PAT via CA-MWCNTs quenched aptamer-tagged carboxyfluorescein. Food Chem. 2020;312:126048. doi: 10.1016/j.foodchem.2019.126048. [DOI] [PubMed] [Google Scholar]
- 131.Esmaelpourfarkhani M., Abnous K., Taghdisi S.M., Chamsaz M. A novel turn-off fluorescent aptasensor for ampicillin detection based on perylenetetracarboxylic acid diimide and gold nanoparticles. Biosens. Bioelectron. 2020;164:112329. doi: 10.1016/j.bios.2020.112329. [DOI] [PubMed] [Google Scholar]
- 132.Wen D., Liu Q., Li L., Yang H., Kong J. Ultrasensitive aptamer fluorometric detection of IFN-γ by dual atom transfer radical polymerization amplification. Sens. Actuators B Chem. 2019;295:40–48. doi: 10.1016/j.snb.2019.05.036. [DOI] [Google Scholar]
- 133.Wang D.-E., Gao X., You S., Chen M., Ren L., Sun W., Yang H., Xu H. Aptamer-functionalized polydiacetylene liposomes act as a fluorescent sensor for sensitive detection of MUC1 and targeted imaging of cancer cells. Sens. Actuators B Chem. 2020;309:127778. doi: 10.1016/j.snb.2020.127778. [DOI] [Google Scholar]
- 134.Fan K., Yang R., Zhao Y., Zang C., Miao X., Qu B., Lu L. A fluorescent aptasensor for sensitive detection of isocarbophos based on AT-rich three-way junctions DNA templated copper nanoparticles and Fe3O4@GO. Sens. Actuators B Chem. 2020;321:128515. doi: 10.1016/j.snb.2020.128515. [DOI] [Google Scholar]
- 135.Wang S.-E., Si S. Aptamer biosensing platform based on carbon nanotube long-range energy transfer for sensitive, selective and multicolor fluorescent heavy metal ion analysis. Anal. Methods. 2013;5:2947–2953. doi: 10.1039/c3ay40360b. [DOI] [Google Scholar]
- 136.Wang M., Hou W., Mi C.-C., Wang W.-X., Xu Z.-R., Teng H.-H., Mao C.-B., Xu S.-K. Immunoassay of Goat Antihuman Immunoglobulin G Antibody Based on Luminescence Resonance Energy Transfer between Near-Infrared Responsive NaYF4:Yb, Er Upconversion Fluorescent Nanoparticles and Gold Nanoparticles. Anal. Chem. 2009;81:8783–8789. doi: 10.1021/ac901808q. [DOI] [PubMed] [Google Scholar]
- 137.Chen Z., Chen H., Hu H., Yu M., Li F., Zhang Q., Zhou Z., Yi T., Huang C. Versatile Synthesis Strategy for Carboxylic Acid−functionalized Upconverting Nanophosphors as Biological Labels. J. Am. Chem. Soc. 2008;130:3023–3029. doi: 10.1021/ja076151k. [DOI] [PubMed] [Google Scholar]
- 138.Huang L., Wu J., Zheng L., Qian H., Xue F., Wu Y., Pan D., Adeloju S.B., Chen W. Rolling Chain Amplification Based Signal-Enhanced Electrochemical Aptasensor for Ultrasensitive Detection of Ochratoxin A. Anal. Chem. 2013;85:10842–10849. doi: 10.1021/ac402228n. [DOI] [PubMed] [Google Scholar]
- 139.Wang F., Liu X. Recent advances in the chemistry of lanthanide-doped upconversion nanocrystals. Chem. Soc. Rev. 2009;38:976–989. doi: 10.1039/b809132n. [DOI] [PubMed] [Google Scholar]
- 140.Wang P., Wang A., Hassan M., Ouyang Q., Li H., Chen Q. A highly sensitive upconversion nanoparticles-WS2 nanosheet sensing platform for Escherichia coli detection. Sens. Actuators B Chem. 2020;320:128434. doi: 10.1016/j.snb.2020.128434. [DOI] [Google Scholar]
- 141.Li H., Ahmad W., Rong Y., Chen Q., Zuo M., Ouyang Q., Guo Z. Designing an aptamer based magnetic and upconversion nanoparticles conjugated fluorescence sensor for screening Escherichia coli in food. Food Control. 2020;107:106761. doi: 10.1016/j.foodcont.2019.106761. [DOI] [Google Scholar]
- 142.Chen Q., Sheng R., Wang P., Ouyang Q., Wang A., Ali S., Zareef M., Hassan M. Ultra-sensitive detection of malathion residues using FRET-based upconversion fluorescence sensor in food. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020;241:118654. doi: 10.1016/j.saa.2020.118654. [DOI] [PubMed] [Google Scholar]
- 143.Chen M., Hassan M., Li H., Chen Q. Fluorometric determination of lead (II) by using aptamer-functionalized upconversion nanoparticles and magnetite-modified gold nanoparticles. Microchim. Acta. 2020;187:85. doi: 10.1007/s00604-019-4030-4. [DOI] [PubMed] [Google Scholar]
- 144.Liu Q., Wang H., Han P., Feng X. Fluorescent aptasensing of chlorpyrifos based on the assembly of cationic conjugated polymer-aggregated gold nanoparticles and luminescent metal–organic frameworks. Analyst. 2019;144:6025–6032. doi: 10.1039/C9AN00943D. [DOI] [PubMed] [Google Scholar]
- 145.Qiu Q., Gao R.-R., Xie A., Jiao Y., Dong W. A ratiometric fluorescent sensor with different DNA-templated Ag NCs as signals for ATP detection. J. Photochem. Photobiol. A Chem. 2020;400:112725. doi: 10.1016/j.jphotochem.2020.112725. [DOI] [Google Scholar]
- 146.Huang L., Wang D.-B., Singh N., Yang F., Gu N., Zhang M. A dual-signal amplification platform for sensitive fluorescence biosensing of leukemia-derived exosomes. Nanoscale. 2018;10:20289–20295. doi: 10.1039/C8NR07720G. [DOI] [PubMed] [Google Scholar]
- 147.Yu X., He L., Pentok M., Yang H., Yang Y., Li Z., He N., Deng Y., Li S., Liu T., et al. An aptamer-based new method for competitive fluorescence detection of exosomes. Nanoscale. 2019;11:15589–15595. doi: 10.1039/C9NR04050A. [DOI] [PubMed] [Google Scholar]
- 148.Huang R., He L., Li S., Liu H., Jin L., Chen Z., Zhao Y., Li Z., Deng Y., He N. A simple fluorescence aptasensor for gastric cancer exosome detection based on branched rolling circle amplification. Nanoscale. 2019;12:2445–2451. doi: 10.1039/C9NR08747H. [DOI] [PubMed] [Google Scholar]
- 149.Gao M.-L., He F., Yin B.-C., Ye B.-C. A dual signal amplification method for exosome detection based on DNA dendrimer self-assembly. Analyst. 2019;144:1995–2002. doi: 10.1039/C8AN02383B. [DOI] [PubMed] [Google Scholar]
- 150.Zhao X., Zhang W., Qiu X., Mei Q., Yang M., Fu W. Rapid and sensitive exosome detection with CRISPR/Cas12a. Anal. Bioanal. Chem. 2020;412:601–609. doi: 10.1007/s00216-019-02211-4. [DOI] [PubMed] [Google Scholar]
- 151.Wang Y., Kang K., Wang S., Kang W., Cheng C., Niu L.M., Guo Z. A novel label-free fluorescence aptasensor for dopamine detection based on an Exonuclease III- and SYBR Green I- aided amplification strategy. Sens. Actuators B Chem. 2020;305:127348. doi: 10.1016/j.snb.2019.127348. [DOI] [Google Scholar]
- 152.Xue N., Wu S., Li Z., Miao X. Ultrasensitive and label-free detection of ATP by using gold nanorods coupled with enzyme assisted target recycling amplification. Anal. Chim. Acta. 2020;1104:117–124. doi: 10.1016/j.aca.2019.12.073. [DOI] [PubMed] [Google Scholar]
- 153.Guo Z., Lv L., Cui C., Wang Y., Ji S., Fang J., Yuan M., Yu H. Detection of aflatoxin B1 with a new label-free fluorescent aptasensor based on exonuclease I and SYBR Gold. Anal. Methods. 2020;12:2928–2933. doi: 10.1039/D0AY00967A. [DOI] [PubMed] [Google Scholar]
- 154.Lu X., Fan Z. RecJf exonuclease-assisted fluorescent self-assembly aptasensor for supersensitive detection of pesticides in food. J. Lumin. 2020;226:117469. doi: 10.1016/j.jlumin.2020.117469. [DOI] [Google Scholar]
- 155.Li B., Liu C., Pan W., Shen J., Guo J., Luo T., Feng J., Situ B., An T., Zhang Y., et al. Facile fluorescent aptasensor using aggregation-induced emission luminogens for exosomal proteins profiling towards liquid biopsy. Biosens. Bioelectron. 2020:112520. doi: 10.1016/j.bios.2020.112520. [DOI] [PubMed] [Google Scholar]
- 156.Zhang J., Shi J., Liu W., Zhang K., Zhao H., Zhang H., Zhangabc Z. A simple, specific and “on-off” type MUC1 fluorescence aptasensor based on exosomes for detection of breast cancer. Sens. Actuators B Chem. 2018;276:552–559. doi: 10.1016/j.snb.2018.08.056. [DOI] [Google Scholar]
- 157.Zhu C., Li L., Wang Z., Irfan M., Qu F. Recent advances of aptasensors for exosomes detection. Biosens. Bioelectron. 2020;160:112213. doi: 10.1016/j.bios.2020.112213. [DOI] [PubMed] [Google Scholar]
- 158.Sapkota K., Dhakal S. FRET-Based Aptasensor for the Selective and Sensitive Detection of Lysozyme. Sensors. 2020;20:914. doi: 10.3390/s20030914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kang B., Park S.V., Soh H.T., Oh S.S. A Dual-Sensing DNA Nanostructure with an Ultrabroad Detection Range. ACS Sens. 2019;4:2802–2808. doi: 10.1021/acssensors.9b01503. [DOI] [PubMed] [Google Scholar]
- 160.Lan Y., Qin G., Wei Y., Dong C., Wang L. Highly sensitive analysis of tetrodotoxin based on free-label fluorescence aptamer sensing system. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019;219:411–418. doi: 10.1016/j.saa.2019.04.068. [DOI] [PubMed] [Google Scholar]
- 161.Li S., Ma X., Pang C., Tian H., Xu Z., Yang Y., Lv D., Ge H. Fluorometric aptasensor for cadmium (II) by using an aptamer-imprinted polymer as the recognition element. Microchim. Acta. 2019;186:823. doi: 10.1007/s00604-019-3886-7. [DOI] [PubMed] [Google Scholar]
- 162.Zhang Y., Lai Y., Teng X., Pu S., Yang Z., Pang P., Wang H., Yang C., Yang W., Barrow C.J. Facile fluorescence strategy for sensitive detection of microcystin-LR based on dsDNA-templated copper nanoclusters. Anal. Methods. 2020;12:1752–1758. doi: 10.1039/C9AY02250C. [DOI] [Google Scholar]
- 163.Sharma R., Akshath U.S., Bhatt P., Raghavarao K.S.M.S. Fluorescent aptaswitch for chloramphenicol detection—Quantification enabled by immobilization of aptamer. Sens. Actuators B Chem. 2019;290:110–117. doi: 10.1016/j.snb.2019.03.093. [DOI] [Google Scholar]
- 164.Mansouri-Majd S., Ghasemi F., Salimi A., Sham T. Transport Properties of a Molybdenum Disulfide and Carbon Dot Nanohybrid Transistor and Its Applications as a Hg2+ Aptasensor. ACS Appl. Electron. Mater. 2020;2:635–645. doi: 10.1021/acsaelm.9b00632. [DOI] [Google Scholar]
- 165.Zhao Y., Cui L., Sun Y., Zheng F., Ke W. Ag/CdO NP-Engineered Magnetic Electrochemical Aptasensor for Prostatic Specific Antigen Detection. ACS Appl. Mater. Interfaces. 2018;11:3474–3481. doi: 10.1021/acsami.8b18887. [DOI] [PubMed] [Google Scholar]
- 166.Adegokea O., Pereira-Barros M.A., Zolotovskaya S., Abdolvand A., Nic Daeid N. Aptamer-based cocaine assay using a nanohybrid composed of ZnS/Ag2Se quantum dots, graphene oxide and gold nanoparticles as a fluorescent probe. Microchim. Acta. 2020;187:104–112. doi: 10.1007/s00604-019-4101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li J., Liu L., Ai Y., Liu Y., Sun H., Liang Q. Self-Polymerized Dopamine-Decorated Au NPs and Coordinated with Fe-MOF as a Dual Binding Sites and Dual Signal-Amplifying Electrochemical Aptasensor for the Detection of CEA. ACS Appl. Mater. Interfaces. 2020;12:5500–5510. doi: 10.1021/acsami.9b19161. [DOI] [PubMed] [Google Scholar]
- 168.Kassahun G., Griveau S., Juillard S., Champavert J., Ringuedé A., Bresson B., Tran Y., Bedioui F., Slim C. Hydrogel Matrix-Grafted Impedimetric Aptasensors for the Detection of Diclofenac. Langmuir. 2020;36:827–836. doi: 10.1021/acs.langmuir.9b02031. [DOI] [PubMed] [Google Scholar]
- 169.Shu Y., Ma W., Ji W., Wei H., Mao L. Aptamer superstructure-based electrochemical biosensor for sensitive detection of ATP in rat brain with in vivo microdialysis. Analyst. 2019;144:1711–1717. doi: 10.1039/c8an02077a. [DOI] [PubMed] [Google Scholar]
- 170.Wang L., Wu A., Wei G. Graphene-based aptasensors: From molecule–interface interactions to sensor design and biomedical diagnostics. Analyst. 2018;143:1526–1543. doi: 10.1039/C8AN00081F. [DOI] [PubMed] [Google Scholar]
- 171.Du Y., Li B., Wei H., Wang Y., Wang E. Multifunctional Label-Free Electrochemical Biosensor Based on an Integrated Aptamer. Anal. Chem. 2008;80:5110–5117. doi: 10.1021/ac800303c. [DOI] [PubMed] [Google Scholar]
- 172.Goud K.Y., Hayat A., Catanante G., Satyanarayana M., Gobi K.V., Marty J.L. An electrochemical aptasensor based on functionalized graphene oxide assisted electrocatalytic signal amplification of methylene blue for aflatoxin B1 detection. Electrochim. Acta. 2017;244:96–103. doi: 10.1016/j.electacta.2017.05.089. [DOI] [Google Scholar]
- 173.Cui L., Lu M., Li Y., Tang B., Zhang C.-Y. A reusable ratiometric electrochemical biosensor on the basis of the binding of methylene blue to DNA with alternating AT base sequence for sensitive detection of adenosine. Biosens. Bioelectron. 2018;102:87–93. doi: 10.1016/j.bios.2017.11.025. [DOI] [PubMed] [Google Scholar]
- 174.Cheng A.K.H., Ge B., Yu H.-Z. Aptamer-Based Biosensors for Label-Free Voltammetric Detection of Lysozyme. Anal. Chem. 2007;79:5158–5164. doi: 10.1021/ac062214q. [DOI] [PubMed] [Google Scholar]
- 175.Gao J., Chen Y., Ji W., Gao Z., Zhang J. Synthesis of a CdS-decorated Eu-MOF nanocomposite for the construction of a self-powered photoelectrochemical aptasensor. Analyst. 2019;144:6617–6624. doi: 10.1039/C9AN01606F. [DOI] [PubMed] [Google Scholar]
- 176.Kwon J., Lee Y., Lee T., Ahn J.-H. Aptamer-Based Field-Effect Transistor for Detection of Avian Influenza Virus in Chicken Serum. Anal. Chem. 2020;92:5524–5531. doi: 10.1021/acs.analchem.0c00348. [DOI] [PubMed] [Google Scholar]
- 177.Wang M., Hu M., Li Z., He L., Song Y., Jia Q., Du M., Du M. Construction of Tb-MOF-on-Fe-MOF conjugate as a novel platform for ultrasensitive detection of carbohydrate antigen 125 and living cancer cells. Biosens. Bioelectron. 2019;142:111536. doi: 10.1016/j.bios.2019.111536. [DOI] [PubMed] [Google Scholar]
- 178.Ran G., Wu F., Ni X., Li X., Li X., Liu D., Sun J., Xie C., Yao D.-S., Bai W. A novel label-free electrochemical aptasensor with one-step assembly process for rapid detection of lead (II) ions. Sens. Actuators B Chem. 2020;320:128326. doi: 10.1016/j.snb.2020.128326. [DOI] [Google Scholar]
- 179.Zhang J., Shang M., Gao Y., Yan J., Song W. High-performance VS2 QDs-based type II heterostructured photoanode for ultrasensitive aptasensing of lysozyme. Sens. Actuators B Chem. 2020;304:127411. doi: 10.1016/j.snb.2019.127411. [DOI] [Google Scholar]
- 180.Chen Y., Xiang J., Liu B., Chen Z., Zuo X. Gold nanoparticle-engineered electrochemical aptamer biosensor for ultrasensitive detection of thrombin. Anal. Methods. 2020;12:3729–3733. doi: 10.1039/D0AY01163K. [DOI] [PubMed] [Google Scholar]
- 181.Zhou J., Cheng K., Chen X., Yang R., Lu M., Ming L., Chen Y., Lin Z., Chen D. Determination of soluble CD44 in serum by using a label-free aptamer based electrochemical impedance biosensor. Analyst. 2019;145:460–465. doi: 10.1039/C9AN01764J. [DOI] [PubMed] [Google Scholar]
- 182.Mohan H.K.S.V., Chee W.K., Li Y., Nayak S., Poh C.L., Thean A.V.-Y. A highly sensitive graphene oxide based label-free capacitive aptasensor for vanillin detection. Mater. Des. 2020;186:108208. doi: 10.1016/j.matdes.2019.108208. [DOI] [Google Scholar]
- 183.Akhtartavan S., Karimi M., Sattarahmady N., Heli H. An electrochemical signal-on apta-cyto-sensor for quantitation of circulating human MDA-MB-231 breast cancer cells by transduction of electro-deposited non-spherical nanoparticles of gold. J. Pharm. Biomed. Anal. 2020;178:112948. doi: 10.1016/j.jpba.2019.112948. [DOI] [PubMed] [Google Scholar]
- 184.Diaz-Amaya S., Lin L.-K., DiNino R.E., Ostos C., Stanciu L. Inkjet printed electrochemical aptasensor for detection of Hg2+ in organic solvents. Electrochim. Acta. 2019;316:33–42. doi: 10.1016/j.electacta.2019.05.079. [DOI] [Google Scholar]
- 185.Ho L.S.J., Fogel R., Limson J. Generation and screening of histamine-specific aptamers for application in a novel impedimetric aptamer-based sensor. Talanta. 2020;208:120474. doi: 10.1016/j.talanta.2019.120474. [DOI] [PubMed] [Google Scholar]
- 186.Song Y., Xu M., Li Z., He L., Hu M., He L., Zhang Z., Du M. Ultrasensitive detection of bisphenol A under diverse environments with an electrochemical aptasensor based on multicomponent AgMo heteronanostructure. Sens. Actuators B Chem. 2020;321:128527. doi: 10.1016/j.snb.2020.128527. [DOI] [Google Scholar]
- 187.Zhang P., Zhang Y., Xiong X., Lu Y., Jia N. A sensitive electrochemiluminescence immunoassay for glycosylated hemoglobin based on Ru(bpy)32+ encapsulated mesoporous polydopamine nanoparticles. Sens. Actuators B Chem. 2020;321:128626. doi: 10.1016/j.snb.2020.128626. [DOI] [Google Scholar]
- 188.Douaki A., Abera B.D., Cantarella G., Shkodra B., Mushtaq A., Ibba P., Inam A.S., Petti L., Lugli P. Flexible Screen Printed Aptasensor for Rapid Detection of Furaneol: A Comparison of CNTs and AgNPs Effect on Aptasensor Performance. Nanomaterials. 2020;10:1167. doi: 10.3390/nano10061167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhao J., Liang D., Gao S., Hu X., Koh K., Chen H. Analyte-resolved magnetoplasmonic nanocomposite to enhance SPR signals and dual recognition strategy for detection of BNP in serum samples. Biosens. Bioelectron. 2019;141:111440. doi: 10.1016/j.bios.2019.111440. [DOI] [PubMed] [Google Scholar]
- 190.Yang Y.-J., Zhou Y., Xing Y., Zhang G.-M., Zhang Y., Zhang C.-H., Lei P., Dong C., Deng X., He Y., et al. A Label-free aptasensor based on Aptamer/NH2 Janus particles for ultrasensitive electrochemical detection of Ochratoxin A. Talanta. 2019;199:310–316. doi: 10.1016/j.talanta.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 191.Song J., Huang M., Jiang N., Zheng S., Mu T., Meng L., Liu Y., Liu J., Chen G. Ultrasensitive detection of amoxicillin by TiO2-g-C3N4@AuNPs impedimetric aptasensor: Fabrication, optimization, and mechanism. J. Hazard. Mater. 2020;391:122024. doi: 10.1016/j.jhazmat.2020.122024. [DOI] [PubMed] [Google Scholar]
- 192.Wang X., Gao F., Gong Y., Liu G., Zhang Y., Ding C. Electrochemical aptasensor based on conductive supramolecular polymer hydrogels for thrombin detection with high selectivity. Talanta. 2019;205:120140. doi: 10.1016/j.talanta.2019.120140. [DOI] [PubMed] [Google Scholar]
- 193.Zhang M., Yin J., Gao C., Qiu G., Meng A., Li Q. The construction of electrochemical aptasensor based on coral-like poly-aniline and Au nano-particles for the sensitive detection of prostate specific antigen. Sens. Actuators B Chem. 2019;295:93–100. doi: 10.1016/j.snb.2019.05.070. [DOI] [Google Scholar]
- 194.Mir M., Vreeke M., Katakis I. Different strategies to develop an electrochemical thrombin aptasensor. Electrochem. Commun. 2006;8:505–511. doi: 10.1016/j.elecom.2005.12.022. [DOI] [Google Scholar]
- 195.Wu Q., Tan R., Mi X., Tu Y. Electrochemiluminescent aptamer-sensor for alpha synuclein oligomer based on a metal–organic framework. Analyst. 2020;145:2159–2167. doi: 10.1039/D0AN00169D. [DOI] [PubMed] [Google Scholar]
- 196.He B., Wang L., Dong X., Yan X., Li M., Yan S., Yan D. Aptamer-based thin film gold electrode modified with gold nanoparticles and carboxylated multi-walled carbon nanotubes for detecting oxytetracycline in chicken samples. Food Chem. 2019;300:125179. doi: 10.1016/j.foodchem.2019.125179. [DOI] [PubMed] [Google Scholar]
- 197.Ding J., Zhang D., Liu Y., Yu M., Zhan X., Zhang D., Zhou P. An electrochemical aptasensor for detection of lead ions using a screen-printed carbon electrode modified with Au/polypyrrole composites and toluidine blue. Anal. Methods. 2019;11:4274–4279. doi: 10.1039/C9AY01256G. [DOI] [Google Scholar]
- 198.Mazaafrianto D.N., Ishida A., Maeki M., Tani H., Tokeshi M. An Electrochemical Sensor Based on Structure Switching of Dithiol-modified Aptamer for Simple Detection of Ochratoxin A. Anal. Sci. 2019;35:1221–1226. doi: 10.2116/analsci.19P240. [DOI] [PubMed] [Google Scholar]
- 199.Wang C., Li Y., Zhao Q. A competitive electrochemical aptamer-based method for aflatoxin B1 detection with signal-off response. Anal. Methods. 2019;12:646–650. doi: 10.1039/C9AY02276G. [DOI] [Google Scholar]
- 200.Ghalehno M.H., Mirzaei M., Torkzadeh-Mahani M. Electrochemical aptasensor for activated protein C using a gold nanoparticle—Chitosan/graphene paste modified carbon paste electrode. Bioelectrochemistry. 2019;130:107322. doi: 10.1016/j.bioelechem.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 201.Fu Y., Callaway Z., Lum J., Wang R., Lin J., Li Y. Exploiting Enzyme Catalysis in Ultra-Low Ion Strength Media for Impedance Biosensing of Avian Influenza Virus Using a Bare Interdigitated Electrode. Anal. Chem. 2014;86:1965–1971. doi: 10.1021/ac402550f. [DOI] [PubMed] [Google Scholar]
- 202.Svigelj R., Dossi N., Pizzolato S., Toniolo R., Miranda-Castro R., De-Los-Santos-Álvarez N., Lobo-Castañón M.J. Truncated aptamers as selective receptors in a gluten sensor supporting direct measurement in a deep eutectic solvent. Biosens. Bioelectron. 2020;165:112339. doi: 10.1016/j.bios.2020.112339. [DOI] [PubMed] [Google Scholar]
- 203.Bai Z., Chen Y., Li F., Yin H., Yin H., Ai S. Electrochemical aptasensor for sulfadimethoxine detection based on the triggered cleavage activity of nuclease P1 by aptamer-target complex. Talanta. 2019;204:409–414. doi: 10.1016/j.talanta.2019.06.035. [DOI] [PubMed] [Google Scholar]
- 204.Xu G., Hou J., Zhao Y., Bao J., Yang M., Fa H., Yang Y., Li L., Huo D., Hou C. Dual-signal aptamer sensor based on polydopamine-gold nanoparticles and exonuclease I for ultrasensitive malathion detection. Sens. Actuators B Chem. 2019;287:428–436. doi: 10.1016/j.snb.2019.01.113. [DOI] [Google Scholar]
- 205.Tan Y., Wei X., Zhang Y., Wang P., Qiu B., Guo L., Lin Z., Yang H.-H. Exonuclease-Catalyzed Target Recycling Amplification and Immobilization-free Electrochemical Aptasensor. Anal. Chem. 2015;87:11826–11831. doi: 10.1021/acs.analchem.5b03314. [DOI] [PubMed] [Google Scholar]
- 206.Sun C., Liu M., Sun H., Lu H., Liu M. Immobilization-free photoelectrochemical aptasensor for environmental pollutants: Design, fabrication and mechanism. Biosens. Bioelectron. 2019;140:111352. doi: 10.1016/j.bios.2019.111352. [DOI] [PubMed] [Google Scholar]
- 207.Zhang R., Wang Y., Qu X., Li S., Zhao Y., Zhang F., Liu S., Huang J., Yu J. A label-free electrochemical platform for the detection of antibiotics based on cascade enzymatic amplification coupled with a split G-quadruplex DNAzyme. Analyst. 2019;144:4995–5002. doi: 10.1039/C9AN00857H. [DOI] [PubMed] [Google Scholar]
- 208.Yi J., Liu Z., Liu J., Liu H., Xia F., Tian D., Zhou C. A label-free electrochemical aptasensor based on 3D porous CS/rGO/GCE for acetamiprid residue detection. Biosens. Bioelectron. 2020;148:111827. doi: 10.1016/j.bios.2019.111827. [DOI] [PubMed] [Google Scholar]
- 209.Wang H., Liu S., Yu J., Xu W., Guo Y., Huang J. Target–aptamer binding triggered quadratic recycling amplification for highly specific and ultrasensitive detection of antibiotics at the attomole level. Chem. Commun. 2015;51:8377–8380. doi: 10.1039/C5CC01473E. [DOI] [PubMed] [Google Scholar]
- 210.Wang W., Xu Y., Cheng N., Xie Y., Huang K., Xu W. Dual-recognition aptazyme-driven DNA nanomachine for two-in-one electrochemical detection of pesticides and heavy metal ions. Sens. Actuators B Chem. 2020;321:128598. doi: 10.1016/j.snb.2020.128598. [DOI] [Google Scholar]