Figure 4:
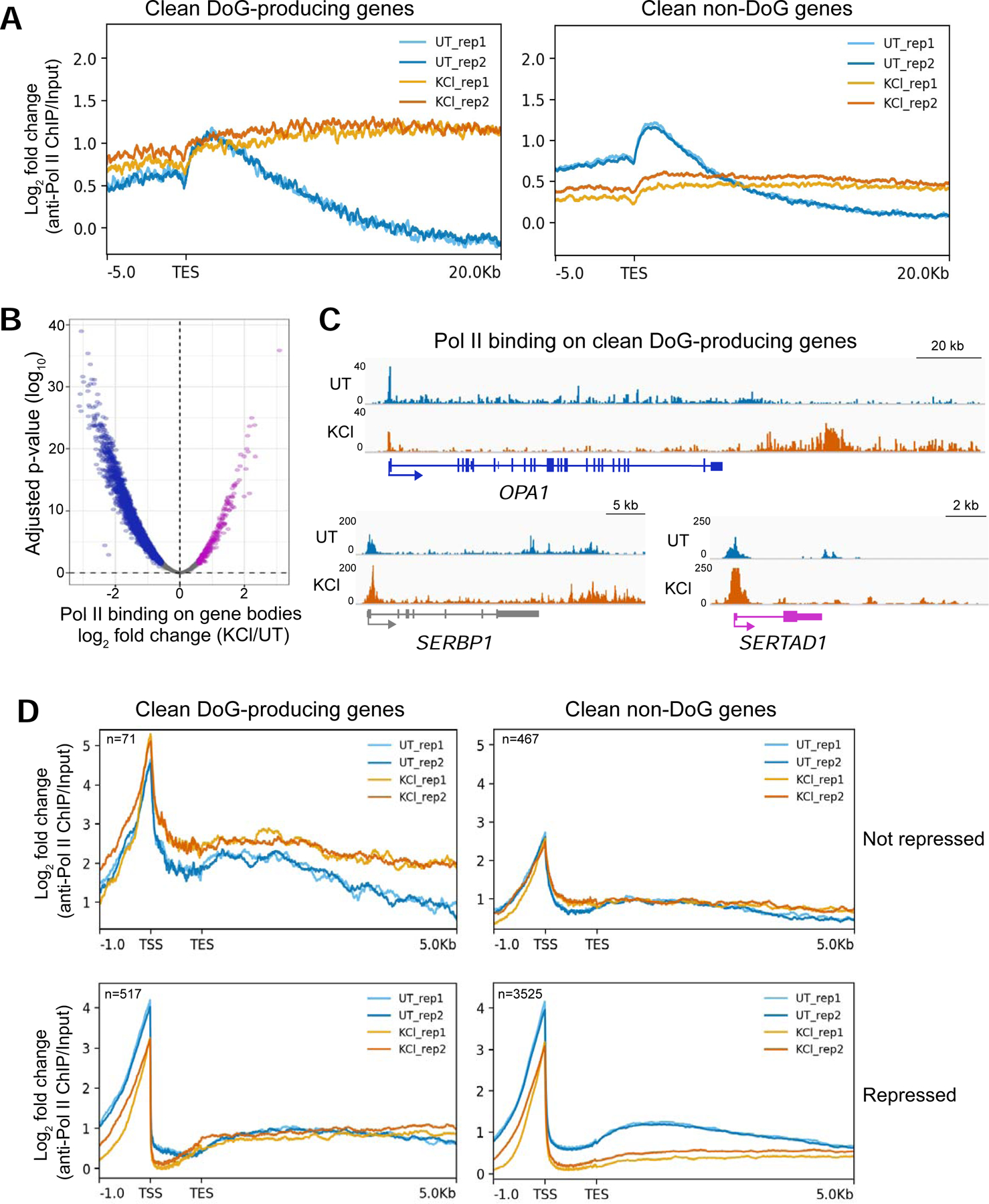
Hyperosmotic stress causes redistribution of Pol II molecules across the genome. A) Meta plots showing the log2 fold change (FC) of anti-Pol II ChIP-seq data normalized to input across the annotated transcription end sites (TES) of DoG-producing clean genes (n=590) and of clean non-DoG genes (n=3994) from UT and KCl-treated HEK-293T cells. B) Volcano plot showing the log2 FC of read counts from anti-Pol II ChIP-seq data normalized to input for clean genes on the x-axis and the corresponding log10 adjusted p-values on the y-axis. Genes with increased Pol II binding across the gene body are shown in purple, genes retaining comparable levels of Pol II binding are gray and those with decreased Pol II binding across the gene body are blue. C) Browser images of DoG-producing clean genes showing anti-Pol II ChIP-seq tracks normalized to input: OPA1 is a repressed gene, SERBP1 retains comparable expression and SERTAD1 is activated by hyperosmotic stress according to TT-TL-seq data. D) Meta plots showing Pol II binding on DoG-producing clean genes and clean non-DoG genes that are not repressed (top) or genes that are repressed (bottom). Two biological replicates are shown in each meta plot for UT and KCl samples.
