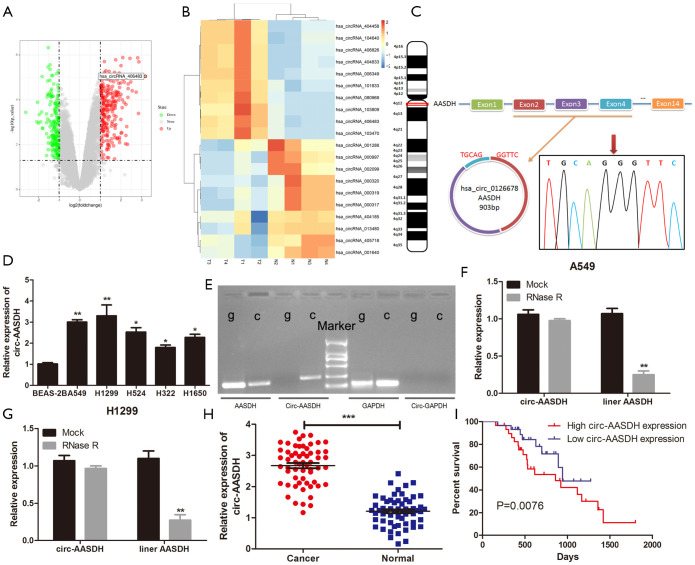
Figure 1.
Circ-AASDH identification and expression in LUAD cells and tissues. (A) The volcano diagram showed the up-regulation and down-regulation of circRNAs in GSE101486. (B) Heatmap come from GEO database (GSE101486) showed the top 20 DEcirc between LUAD samples and normal tissues. (C) Circ-AASDH is formed by cyclization of exon 2–4 of AASDH from chromosome 4. RT-PCR and Sanger sequencing confirmed the "head-tail" splice site (red arrow) of circ-AASDH. (D) Circ-AASDH Expression in LUAD cell lines (A549, H1299, H524, H322, H1650) and normal pulmonary epithelial cell line BEAS-2B. (E) RT-PCR confirmed the existence of circ-AASDH in cDNA. The divergent primers amplified circ-AASDH in cDNA, while didn’t in gDNA. (F,G) The expression levels of circular and linear AASDH in A549 and H1299 cells treated with RNase R were detected by qRT-PCR. (H) Circ-AASDH expression in 59 pairs of LUAD and paracancerous tissues. (I) The OS rates of LUAD patients with high and low expression of circ-AASDH was evaluated by Kaplan-Meier method. LUAD, lung adenocarcinoma. Decirc, differentially expressed circRNA. OS, overall survival. *P<0.05, **P<0.01, ***P<0.001.