Abstract
Background
In advanced non-small cell lung cancer (NSCLC) a recent meta-analysis confirms circulating tumour cells (CTCs) as an independent prognostic indicator of progression-free survival (PFS) and overall survival (OS). However, further investigations are necessary to predict and dynamically monitor the therapy in NSCLC patients using CTCs. To this aim, we combined the classical standard assay (SA) with an expanded cytokeratins profile (EA) and quantified the expression of EML4-ALK fusion protein in CTCs.
Methods
The CellSearch (CS) platform—first marked in vitro diagnostic use (IVD) from Food and Drug Administration (FDA), and “gold standard” for quantifying CTCs - detects EpCAM and cytokeratins (CKs) 8, 18, and 19. Since NSCLC shows different CKs profile, we customized the SA, to recognize CK 4, 5, 6, 7, 8, 10, 13, 14, 18, and 19 (EA). Using both assays we designed a prospective, multi-center study, primarily aimed to enumerate CTCs in advanced NSCLC. Secondarily, we developed an integration of the EA to quantify the expression of EML4-ALK fusion protein in CTCs, and correlated them with PFS and OS.
Results
EA identified a significantly much more number of CTC-positive patients (115 out of 180) than SA (103 out of 192; Chi-square =4.0179, with 1 degrees of freedom, P=0.04502). Similar to SA, EA levels were still associated with patient’ outcomes. Furthermore, the expression of EML4-ALK on CTCs allowed stratifying NSCLC patients according to a statistically significant difference in PFS.
Conclusions
We proposed here two novel automated tests, to characterize the expression of specific molecules on CTCs. We demonstrated that these integrated assays are robust and actionable in prospective clinical studies, and in the future could allow clinicians to improve both choice and length of treatment in individual NSCLC patient.
Keywords: Non-small cell lung cancer (NSCLC), circulating tumour cells (CTCs), EML4-ALK, cytokeratins (CK), liquid biopsy
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide (1).
At diagnosis, more than half of patients are in III/IV stage of disease (2) and biopsies are often insufficient in quantity and quality for molecular analysis or unavailable. Thus, in the last decade tumour cells circulating in the peripheral blood (CTCs) have been extensively investigated as potential disease marker; notably, in non-small cell lung cancer (NSCLC) CTCs can be detected at all disease stages (3).
Two key reports (4,5) used CellSearch (CS) platform that first obtained the IVD mark from FDA, and to date, represents the “gold standard” (6). The patients’ percentage with at least 1 (7) or 2 or more CTCs in 7.5 ml of peripheral blood (PB) (4) varied from 21% and 30,6%, depending on the study. Moreover, CTCs have been proposed as surrogate marker of distant metastasis in the early stages of lung cancer (7).
Several authors then addressed the prognostic significance of CTC enumeration by CS, in NSCLC. A first meta-analysis included 27 articles (12 containing survival outcomes) (8), and concluded that both pre- and post-treatment CTC detections in PB were associated with poor prognosis.
In a second meta-analysis, that examined 20 studies enrolling more than 1,500 NSCLC patients (9), CTCs were associated with tumour stage and lymph-node metastasis, but not with histology; in addition, CTCs were associated with a poorer outcome (9). Recently, EPAC meta-analysis confirms CTCs as an independent prognostic indicator of progression-free survival (PFS) and overall survival (OS) in advanced NSCLC (10).
Some studies also analysed changes of CTC levels as predictor of treatment effectiveness, even if for small samples size, showing that CTCs decreased at the end of the first cycle of therapy in patients with more favourable PFS and OS (11,12). However, a consensus on the predictive value of CTCs is currently lacking. Indeed, the considerable heterogeneity within and between lung cancer patients is a matter of debate, in particular regarding epithelial and mesenchymal antigens expressed in CTCs and about methods used to detect them (13).
In this regard, the main limit on using the standard CS assay (SA) in NSCLC, is that SA recognizes cytokeratins (CKs) 8, 18, and 19, as part of the criteria to define an event as CTC; however, these CKs are not always expressed in lung cancer, where a different CKs profile could be detected (14).
Second, in NSCLC the expression of full-length EpCAM is lower than in other carcinomas (15,16). Likewise, the expression of EpCAM and CKs is heterogeneous in CTCs of stage IV NSCLC (17). Overall, it raises some doubt that CS better fits with lung cancer patients.
To address this item, in the last years huge number of studies and alternative techniques have been emerging, which employ e.g. label-independent, size-based strategies of CTC enrichment (18,19), and allow identifying potential targets of specific treatment (20-22). However, novel procedures, often with different levels of validation (6), produced controversial results and limited transfer from benchtop to bedside (23).
Nevertheless, beyond enumerating, the most intriguing use of CTCs in clinical, could be to identify patients, which show activating mutations, e.g. of epidermal growth factor receptor (EGFR), (17,24), or fusion genes, such as EML4-ALK (25), or ALK-ROS1 (26). In fact, to support clinicians to identify which patients could benefit a targeted drug throughout the continuum of the care, CTCs are an option when tumour biopsy is unavailable or of unacceptable discomfort for the patient.
Although cell-free circulating tumour DNA (ctDNA) can be exploited to this purpose, as the sum of different neoplastic lesions present at any time in individual patient, we previously reported that EGFR mutation could be detected in EpCAM-positive tumour cells, but not in ctDNA (24). Since few mutated CTCs did not affect the total amount of ctDNA to result in EGFR detection, we demonstrated that DNA extracted from CTCs (ctcDNA) investigation was more useful at that time (24). More generally, we reported that quantifying “druggable” CTCs allows predicting treatment effectiveness (27-29).
Therefore, in order to get actionable CTC enumeration in any histotype of NSCLC, including those that lack CK 8, 18 and 19, we integrated the in vitro diagnostic use (IVD) assay (standard assay, SA) with monoclonal antibodies (mAbs) that recognize CK 4, 5, 6, 7, 10, 13, 14; we named this test expanded assay (EA), to stress that it recognizes much more CKs than the SA.
We were aware that choosing an EpCAM-based platform as CS, which enriches and quantifies a pre-defined phenotype of CTCs, we could exclude from our analysis some subsets of CTCs, with eventually higher metastatic capabilities (30). However, since it is not conceivable that one test fit all CTC subpopulations, we focused on implementing in clinical robust automated assays (6), in the view to address the urgent need for real time monitoring, stratification, and personalized treatment of lung cancer patients. Indeed, we previously reported that EpCAMhigh CTCs strongly correlated with poor OS, whilst EpCAMlow CTCs did not (31).
Here we present the results of a prospective, multi-center study, primarily aimed to enumerate CTCs in advanced NSCLC.
The secondary end-points aimed: (I) to correlate CTC enumeration, according to SA and EA, with progression free survival (PFS) and overall survival (OS); (II) to establish a cut-off value of prognostic significance for SA and EA; (III) to quantify the fraction of apoptotic CTC (M30-positive CTCs) (32) and (IV) the expression of EML4-ALK fusion protein in CTCs (25,26), for correlating their levels with PFS and OS.
The authors present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-855).
Methods
Patient cohort
Between December 2012 and June 2016, we enrolled 199 NSCLC patients (GR cohort) coming from 6 Italian clinical sites, namely CROB (Rionero In Vulture), Ospedale Sant’Andrea di Roma (Roma), Campus Biomedico (Roma), IRST (Meldola), IOV-IRCCS (Padova), Azienda Ospedaliera di Perugia (Perugia).
Peripheral blood (PB) samples were collected upon signed informed consent [IOV-IRCCS study “Individualized treatment of patients with advanced NSCLC: potential application for Circulating Tumour Cells (CTCs) molecular and phenotypical profiling”, NCT02407327] and analysed for CTC enumeration in a multi-center prospective observational protocol, headed by the CTC-lab of IOV-IRCCS, and approved by IOV-IRCCS Ethical Committee (protocol # CE IOV 2012/52). Clinicians were blinded on CTC results and the study outcomes did not modify the subsequent management of the patients.
Inclusion criteria were as following:
❖ Age 18–90 years old, male or female;
❖ Patients with advanced NSCLC, confirmed (if allowed by patient’s conditions) by histopathology or cytology;
❖ Life expectancy >6 months.
❖ Any first line therapy.
Exclusion criteria included conditions hampering compliance to assessments schedule. We collected serial blood samples at baseline (T0), at the end of first cycle of therapy (T1), and at the first and subsequent radiological assessments and/or at progression (T2 and T3, respectively).
After baseline evaluation, the treating physicians conducted re-evaluations of the disease status based on Response Evaluation Criteria in Solid Tumour (RECIST).
The T0 CTC enumeration of the GR cohort entered in the European collaborative study of advanced NSCLC, elsewhere published (10).
Whole blood collection for CTC enumeration
Two blood samples (7.5 mL/sample) from each patient were collected into CellSave tubes (code # 790005, Menarini, Italy). We maintained and shipped blood samples at room temperature, processing them upon arrival, within a maximum of 96 h after collection.
CTC detection by CellSearch
Investigation of cytokeratins profile
We enumerated EpCAM-positive CTCs in whole blood with the CellSearch System (CS) (Menarini, Italy) according to the manufacturer’s instructions of the Circulating Tumour Cell (CTC) Kit (code # 7900001, Menarini, Italy) and user’s guidelines (IVD standard assay, SA) (33,34).
Furthermore, to investigate the pan-cytokeratins (CK) profile of NSCLC CTCs, (expanded assay, EA) we customized the standard test, as previously described, using the CXC Kit (code # 7900018, Menarini, Italy) in conjunction with FITC-conjugated mAbs anti-CK7, -CK14 (clone LP5K and LL002, respectively, Millipore, Billerica MA, USA), and anti-CK 4, 5, 6, 8, 10, 13, 18, and 19 (clone C11, ACZON, Bologna, Italy) (24).
Detection of apoptosis level
Moreover, we quantify the fraction of apoptotic CTCs, using the specific mAb, M30 CytoDEATH Fluorescein (ALX-804-590, Alexis Biochemicals) in the extra marker channel of SA (32). We expressed results as total number of CTC and M30-positive CTC per 7.5 mL of PB.
Detection of EML4-ALK fusion protein
In parallel, we investigated the expression of EML4-ALK fusion protein, by adding to the 4th custom channel of the EA assay, the mAb anti-ALK–PE (clone D5F3 CellSignaling) (25), validated to be used in IHC of primary tumour.
The clone D5F3 was selected since in highly expressing (3+) cases, it shows sufficiently high correlations with presence of an EML4-ALK fusion transcript, to be used in clinical practice.
Hence, we performed FISH analysis in negative control—H460 cell line—and positive (3+) control—A549 cell line. The human cell lines H460 and A549 have been obtained from ATCC.
We then compared FISH results and EML4-ALK images obtained by CS with the same cell lines; these were then used as reference, when we applied the EA integrated for EML4-ALK expression in NSCLC patients (see Figure S1 and Figure S2).
We expressed results as the total number of CTCs and EML4-ALK-positive CTCs per 7.5 mL of PB.
A schematic overview of the four CS assays exploited in the study is reported in Supplementary materials.
Analysis of ALK rearrangement by fluorescence in situ hybridization (FISH)
ALK rearrangement was evaluated by fluorescence in situ hybridization (FISH) in biopsy specimens, and in the human cell lines H460 and A549.
Briefly, specimens were formalin-fixed and paraffin-embedded (FFPE), according to the manufacturer’s instructions (DakoCytomation, Glostrup, Denmark). FFPE 4-µm-thick sections were mantained at 60 °C for 60 min, deparaffinized and rehydrated. After pretreatment solution—10 min at 95 °C—samples were digested with pepsin solution for 8 min at room temperature. Dual-color, break-apart probe specific for the ALK locus on chromosome 2p23 (ALK FISH DNA Probe, Split Signal, DakoCytomation, Glostrup, Denmark) was hybridized according to the manufacturer’s instructions. Post-hybridization stringency wash was performed in water bath at 65 °C for 10 min. After washing twice and drying at room temperature for 15 min, slides were mounted with fluorescent mounting medium containing 4’6-diamidine-2-phenylindole (DAPI, DakoCytomation, Glostrup, Denmark). A microscope Nikon Eclipse 80i with single and triple band-pass filters was employed to detect FISH signals. The Genikon System was used to acquire and process images. At least 100 tumour cells were scored for the presence of split signals. A case was considered positive for ALK rearrangement if >15% of cells showed split signals.
Statistical analysis
The association of CTCs with individual clinical characteristics, including stage, Performance Status (PS), histology, smoking status, and sites of metastasis, was compared by Chi-square test or Fisher’s exact test, where appropriate.
Quantitative variables in different groups were compared by Mann-Whitney rank sum, non-parametric test.
PFS was measured from date of baseline blood sample to date of confirmed clinical progression or death or was censored at last follow-up. OS was measured from date of baseline blood sample to date of death or was censored at last follow-up.
Kaplan-Meier curve method was used to estimate survivor functions and the Log-rank test was performed to test the differences between curves. Unadjusted Cox regression model was then employed to estimate the effect of CTC enumeration by SA and EA on OS and on PFS through the hazard rate ratio (RR) (and its 95% confidence interval). Cox model were then adjusted by sex, age, smoking status and performing status.
Descriptive statistical analysis was performed using SPSS version 13.0.2004 (SPSS, Chicago, IL); Kaplan-Meier curve and Cox models were analysed using SAS v. 9.4 using PROC LIFETEST and PROC PHREG.
Results
Patient demographics
Between December 2012 and June 2016, we consecutively enrolled 199 patients with advanced NSCLC (GR cohort). Table 1 summarizes the demographic characteristics of GR cohort.
Table 1. Demographic characteristics of GR cohort.
| Patients | N=199 |
|---|---|
| Age, years | |
| Mean ± SD | 64.52±10.9 |
| Median [range] | 65 [28–86] |
| Gender, n (%) | |
| Male | 122 (61.3) |
| Female | 77 (38.7) |
| Performance Status at baseline CTC run (ECOG), n (%) | |
| 0 | 36 (18.1) |
| 1 | 97 (48.7) |
| 2 | 25 (12.6) |
| 3 | 10 (5.0) |
| 4 | 3 (1.5) |
| Not specified | 28 (14.1) |
| Stage, n (%) | |
| Stage IIIb | 6 (3.0) |
| Stage IV | 146 (73.3) |
| Not specified | 47 (23.6) |
| NSCLC histology, n (%) | |
| Adenocarcinoma | 174 (87.4) |
| Squamous | 16 (8.0) |
| Other | 7 (3.6) |
| Not determined | 2 (1.0) |
| Smoking status, n (%) | |
| Never | 37 (18.6) |
| Ex | 71 (35.7) |
| Current | 40 (20.1) |
| Not specified | 51 (25.6) |
The table shows the number (2nd column) and percentage (between round brackets) of patients having specific clinical pathological characteristics.
Prevalence of CTCs using SA and EA at baseline
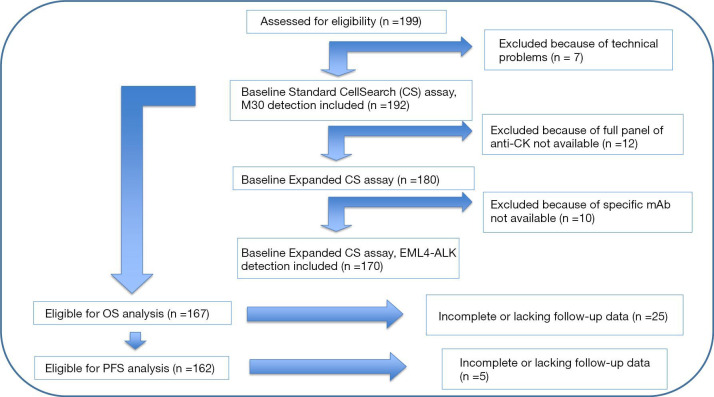
The Consort diagram (Figure 1) describes study assessments.
Figure 1.
Consort diagram. According to manufacturer guidelines, we excluded 7 patients from the analysis for technical problems, since their samples did not satisfy pre-analytical quality criteria (micro-clotting, haemolysis, sample delivery beyond 96 hours from the blood draw). Moreover, 12 patients were excluded from EA since the full panel of anti-CK antibodies was not available and a further group of 10 patients because the specific antibody for EML4-ALK detection was not available at the time of their enrolment.
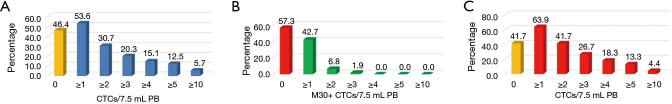
Using SA, CTCs ranged from 0 to 100 cells [median 1 cells, inter quartile range (IQR) 3]. More specifically, 103 out of 192 NSCLC patients (53.6%) had at least 1 CTC in 7.5 mL of PB (Figure 2A), whilst 59 out of 192 (30.7%) had 2 or more CTCs.
Figure 2.
Prevalence of circulating tumor cell (CTC) numbers, using different tests. (A) The picture shows the percentage of GR cohort (n=192) that had <1 CTC/7.5 mL PB (yellow histogram) or increasing number of CTCs (blue histograms), using SA; (B) the picture shows the percentage of CTC-positive patients by SA (n=103) that had <1 M30-positive CTC (red histogram) or increasing number of M30-positive CTCs (green histograms); (C) the picture shows the percentage of GR cohort (n=180) that had <1 CTC/7.5 mL PB (yellow histogram) or increasing number of CTCs (red histograms), using the EA.
Interestingly, this last frequency did not statistically differ from data previously reported in a cohort of only 60 stage IV NSCLC (59 out of 192 vs. 19 out of 60 patients; Chi-square =0.00188, with 1 degrees of freedom, P=0.891) (5). Conversely, CTCs at baseline did not associate with distant metastasis (lung, mediastinal LN, liver or bone; not shown) as previously reported by other studies that included NSCLC patients of stages III and IV (5).
Since apoptotic CTCs has been inversely associated to their metastatic capability, we then evaluated the M30 expression in CTC-positive patients. We found that 44 out of 103 cases (42.7%) had 1 or more M30-positive CTCs (Figure 2B). The M30-positive CTCs ranged from 0 to 3 cells (median 0). Overall, the percentage of CTC-positive patients that showed at least 1 M30-positive CTC was lower than in other malignancies (32). CTCs and M30-positive CTCs at diagnosis did not associate with any specific clinic-pathologic features.
Using the EA, the median number of CTCs was still 1 cell, and the IQR was 3.
However, using the EA assay we found that 115 patients out of 180 (63.9%) had at least 1 CTC in 7.5 mL of PB, whilst 75 out of 180 (41.7%) patients had 2 or more CTCs (Figure 2C).
Notably, the EA allows revealing a number of CTC-positive patients (115 out of 180) significantly higher than SA (103 out of 192; Chi-square =4.0179, with 1 degrees of freedom, P=0.04502).
We then evaluated the correlation of CTC level with clinical and demographic characteristic. The prevalence of CTC-positive patients (Table S1) was higher in women who have NSCLC compared to men according to SA (Chi-square =4.7147 with 1 degrees of freedom, P=0.0299) but not with EA (Chi-square =0.3742 with 1 degrees of freedom, P=0.5407).
Moreover, using EA the prevalence of CTC-positive patients resulted higher in never smoking patients (Chi-square =7.0017 with 2 degrees of freedom, P=0.030171).
To further investigate this finding, we then analysed the group of 78 out of 84 CTC-positive patients, which had both EA and EML4-ALK analysis (Table S2).
We found no significant differences according to smoking habits, (Chi-square statistic =0.8905 with 2 degree of freedom, P=0.640661) in the frequency of EML4-ALK expression in CTC-positive patients (n=78), whilst a significant higher prevalence of never-smoking women was found in this group (Chi-square statistic =12.1668 with 2 degree of freedom, P=0.00228).
The frequencies of patients that had at least 1 EML4-ALK-positive CTCs (ALK+, n=25) were then evaluated according to smoking habits and gender (Table S2).
Notably, EA revealed a significant higher prevalence of ALK+ cases [at least 1 EML4-ALK-positive CTC/7.5 mL PB (n=25)] in never-smoking women (Fisher’s Exact test, P=0.00233).
PFS according to SA and EA at baseline
To evaluate SA and EA levels as prognosticator at baseline in GR cohort, we analysed the PFS that includes death or progression during or after the treatment scheduled after the T0 pre-treatment CTC enumeration.
CTCs always resulted associated with a poorer prognosis. In fact, according to SA test (Figure 3A), PFS of CTC-positive patients was significantly lower than of CTC-negative (0.46 and 0.94 years, respectively; Kaplan-Meyer, Log Rank test P=0.002). Similarly, according to EA results, patients with 4 or more CTCs had a significantly lower median PFS compared to patients, which had less than 4 CTCs (0.29 vs. 0.66 years; Kaplan-Meyer, Log-Rank test P=0.004) (Figure 3B).
Figure 3.
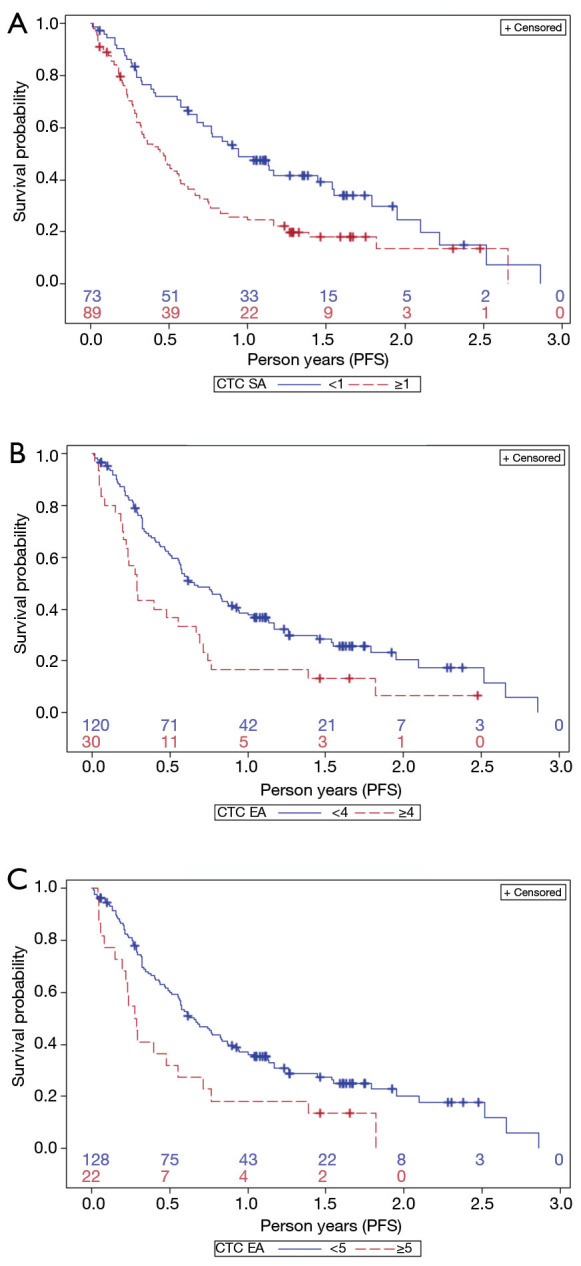
Prediction of progression-free survival (PFS) based on circulating tumour cell (CTC) count as determined by SA (A) using 1 CTC as cut-off. Prediction of PFS using EA with 4 CTCs (B) and 5 CTCs (C) as cut-off value. (A) Kaplan-Meir curves identify two groups of patients with favourable (blue line) or unfavourable (dotted red line) prognosis, depending on CTC <1 or ≥1 cells/7.5 mL PB, enumerated by SA (Log Rank test P=0.002); (B) Kaplan-Meir curves identify two groups of patients with favourable or unfavourable prognosis, depending on CTCs <4 or ≥4 cells/7.5 mL PB, enumerated by EA (Log Rank test P=0.004); (C) Kaplan-Meir curves identify two groups of patients with favourable or unfavourable prognosis, depending on CTCs <5 or ≥5 cells/7.5 mL PB, enumerated by EA (Log Rank test P=0.007).
Similar results hold for 5 or more CTCs (0.28 vs. 0.66 years, P value =0.007) (Figure 3C), and 6 or more CTCs (0.29 vs. 0.63, P value =0.01).
The unadjusted Cox regression model estimated the effect of SA and EA (for all different cut-off) on PFS (Table 2). Using SA, CTC-negative patients have 0.56 times the risk of PFS (RR =0.56; 95% CI: 0.39–0.81) compared to CTC-positive patients (i.e., CTC-negative patients have 44% less risk of PFS). This finding still held after adjusting for sex, age, smoking status, performance status (RR =0.51; 95% CI: 0.33–0.79).
Table 2. Survival median and hazard rate ratio (95% CI) for progression free survival and overall survival, by different cut-offs.
| PFS | OS | ||||||
|---|---|---|---|---|---|---|---|
| Kaplan-Meier | Cox-unadjusted | Cox-adjusted* | Kaplan-Meier | Cox-unadjusted | Cox-adjusted* | ||
| SA | |||||||
| Negative vs. positive | n=162 | n=162 | n=119 | n=167 | n=167 | n=123 | |
| Median =0.94 vs. 0.46 | RR =0.56 | RR =0.51 | Median =1.62 vs. 0.83 | RR =0.58 | RR =0.51 | ||
| P value =0.002 | (0.39–0.81) | (0.33–0.79) | P value=0.007 | (0.39–0.86) | (0.31–0.81) | ||
| EA | |||||||
| Negative vs. positive | n=150 | n=150 | n=113 | n=154 | n=154 | n=116 | |
| Median =0.83 vs. 0.49 | RR =0.76 | RR =0.68 | Median =1.18 vs. 0.90 | RR =0.78 | RR =0.82 | ||
| P value=0.17 | (0.51–1.13) | (0.43–1.09) | P value =0.27 | (0.50–1.21) | (0.49–1.38) | ||
| <2 vs. ≥2 | n=150 | n=150 | n=113 | n=154 | n=134 | n=116 | |
| Median =0.77 vs. 0.36 | RR =0.61 | RR =0.77 | Median =1.29 vs. 0.86 | RR =0.71 | RR =0.91 | ||
| P value=0.001 | (0.42–0.89) | (0.49–1.20) | P value=0.08 | (0.47–1.08) | (0.59–1.40) | ||
| <3 vs. ≥3 | n=150 | n=150 | n=113 | n=154 | n=134 | n=116 | |
| Median =0.61 vs. 0.43 | RR =0.79 | RR =0.89 | Median =1.19 vs. 0.84 | RR =0.82 | RR =0.99 | ||
| P value =0.25 | (0.53–1.18) | (0.55–1.42) | P value=0.40 | (0.53–1.28) | (0.63–1.57) | ||
| <4 vs. ≥4 | n=150 | n=150 | n=113 | n=154 | n=134 | n=116 | |
| Median =0.66 vs. 0.29 | RR =0.54 | RR =0.62 | Median =1.29 vs. 0.59 | RR =0.61 | RR =0.90 | ||
| P value =0.004 | (0.35–0.83) | (0.36–1.06) | P value=0.04 | (0.38–0.99) | (0.53–1.50) | ||
| <5 vs. ≥5 | n=150 | n=150 | n=113 | n=154 | n=134 | n=116 | |
| Median =0.66 vs. 0.28 | RR =0.52 | RR =0.64 | Median =1.25 vs. 0.49 | RR =0.49 | RR =0.62 | ||
| P value =0.007 | (0.32–0.84) | (0.35–1.15) | P value =0.008 | (0.29–0.82) | (0.36–1.06) | ||
| <6 vs. ≥6 | n=150 | n=150 | n=113 | n=154 | n=134 | n=116 | |
| Median =0.63 vs. 0.29 | RR =0.52 | RR= 0.55 | Median =1.19 vs. 0.59 | RR =0.50 | RR =0.63 | ||
| P value =0.01 | (0.31–0.7) | (0.29–1.04) | P value =0.022 | (0.29–0.88) | (0.36–1.12) | ||
*, Cox regression analysis has been adjusted by sex/age/smoking/performance status. Kaplan-Meyer, Cox Regression unadjusted and Cox Regression adjusted analyses of PFS and OS were used for CTC level considered as a binary (<1 or ≥1 CTC/7.5 mL PB), i.e., negative vs. positive, according to SA enumeration. Different cut-off values were considered for CTC enumeration by EA, from <1 vs. ≥1, up to <6 vs. ≥6 CTCs/7.5 mL PB.
Using EA, the patients with less than 2 CTCs have a 0.61-times (i.e., a 39% decreased in risk) the risk of PFS (RR =0.61; 95% CI: 0.42–0.89) compared to patients with 2 or more CTCs, at the unadjusted level of analysis. Similar results hold for all other cut-offs but for less than 3 CTCs.
However, after adjusting by sex, age, smoking status and performance status, the CTC level as measured by EA (for all cut-off) did not show association with PFS. We cannot exclude that this is due to lower sample size available for adjusted analysis (n=113) vs. the unadjusted one (n=150) (Table 2).
Overall survival (OS) according to SA and EA at baseline
Overall, the median OS of the GR cohort (n=199) was 198 days. According to SA, the CTC-negative patients showed a median OS of 1.62 years significantly higher than 0.83 years for the CTC-positive patients (Kaplan-Meier, Log-rank test, P=0.007) (Figure 4A).
Figure 4.
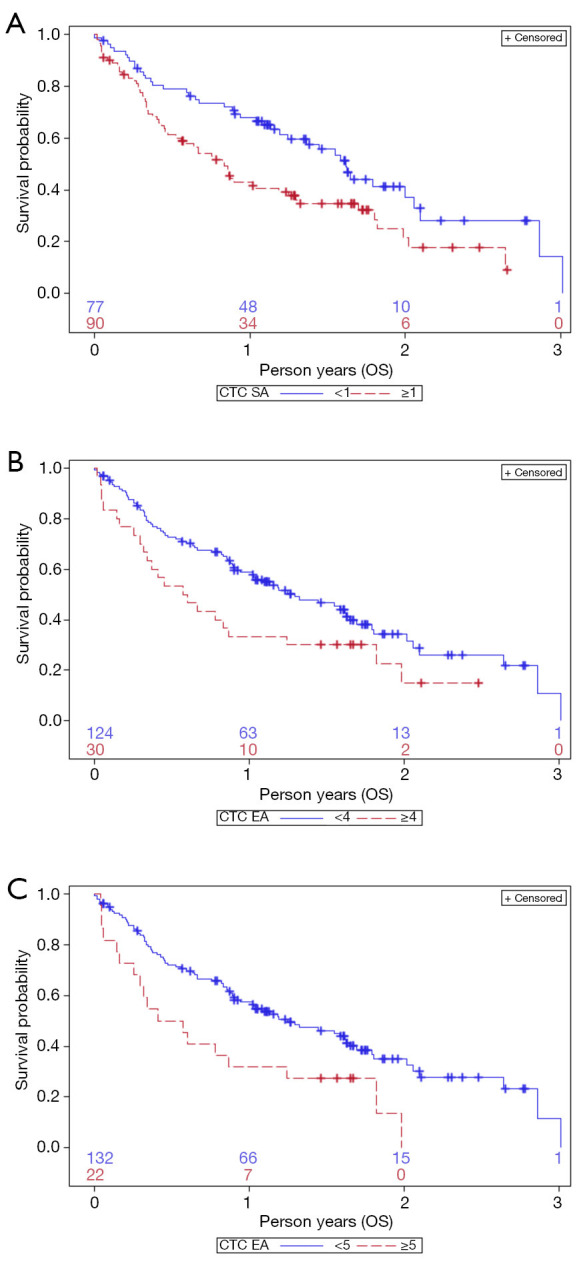
Prediction of overall survival (OS) based on circulating tumour cell (CTC) count as determined by SA (A) using 1 CTC as cut-off. Prediction of OS analysed with EA using 4 CTCs (B) and 5 CTCs (C) as cut-off value. (A) Kaplan-Meir curves identify two groups of patients with favourable (blue line) or unfavourable (dotted red line) prognosis, depending on CTC <1 or ≥1 cells/7.5 mL PB, enumerated by SA (Log Rank test P=0.007); (B) Kaplan-Meir curves identify two groups of patients with favourable or unfavourable prognosis, depending on CTCs <4 or ≥4 cells/7.5 mL PB, enumerated by EA (Log Rank test P=0.04); (C) Kaplan-Meir curves identify two groups of patients with favourable or unfavourable prognosis, depending on CTCs <5 or ≥5 cells/7.5 mL PB, enumerated by EA (Log Rank test P=0.008).
Moreover, when we used 4 CTCs as cut-off value for EA, patients with CTCs <4 showed a median OS of 1.29 years, significantly longer than in patients with CTC ≥4, which showed a median OS of 0.59 years (Long-Rank test P=0.04) (Figure 4B). A higher cut-off value (CTC ≥5) further increased the significance level of Log-rank test (P=0.008, Figure 4C), since it further reduced the median OS of the high-risk group to 0.49 years, whereas the lower-risk group (CTC <5) had a median OS of 1.25 years.
The unadjusted Cox regression model estimated the effect of SA and EA (for all different cut-off) on OS (Table 2). Using SA, the CTC-negative patients resulted to have a 0.58-times the risk of OS (i.e., a 42% decreased risk) compared to CTC-positive patients (RR =0.58; 95% CI: 0.39–0.86). This finding still held after adjusting for sex, age, smoking status, performance status (RR =0.51; 95% CI: 0.31–0.81).
Using EA, the CTC level showed a decreased risk at the unadjusted level of analysis, with <4 cells as cut-off (RR =0.61; 95% CI: 0.38–0.99), with <5 cells as cut-off (RR =0.49; 95% CI: 0.29–0.82), and with <6 cells as cut-off (RR=0.50; 95% CI: 0.29–0.88); but, when adjusting by sex, age, smoking status and performance status the RR was not significant anymore.
We cannot exclude that this is due to lower sample size available for adjusted analysis (n=134) vs. the unadjusted one (n=154) (Table 2).
CTCs characterization: EML4-ALK expression
To determine the expression of the fusion protein EML4-ALK on CTCs we developed an integration of the EA, as previously reported for other customized assay (32).
At first, the anti-ALK–PE (clone D5F3) was used in conjunction with EA, to determine the EM4-ALK expression of two human lung cancer cell lines, H460 and A549, which served as negative and positive control, respectively. The selection of these cell lines has been based on FISH analysis that showed negative signal in H460 cell line, and positive signal (3+) in A549 cell line (Figure S1A).
To be processed by CS platform, H460 and A549 cells were spiked into whole blood sample of healthy donor, at concentration of 300 cells/7.5 mL, i.e., in the range of the maximum level observed in-vivo in cancer patients (200–1,000 cells/7.5 mL peripheral blood).
The analysis of spiked-samples allowed determining the compatibility of EML4-ALK immunostaining with the EA assay in appropriate controls, and to optimize the exposure/integration time (IT) of the image scanning (Figure S1B). The EML4-ALK integrated assay has been then fully developed in PB specimens obtained from NSCLC patients (Figure S2).
We then applied the EML4-ALK-CTC assay to the GR cohort.
At baseline, we found at least 1 EML4-ALK-positive CTC in 35 out 105 patients (33.3%) (Figure 5A). This percentage is higher than 3–7% previously reported in NSCLC biopsies (35,36).
Figure 5.
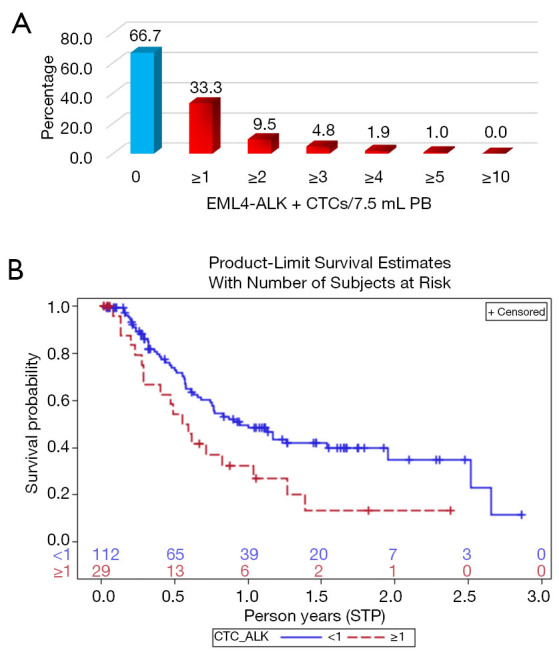
Prevalence of EML4-ALK-positive circulating tumour cells (CTCs) and prediction of progression-free survival (PFS) based on EML4-ALK expression on CTCs. (A) The picture shows the percentage of CTC-positive patients by EA (n=105) that had <1 EML4-ALK-positive CTC (blue histogram) or increasing number of EML4-ALK-positive CTCs (red histograms); (B) Kaplan-Meir curves identify two groups of patients with favourable (blue line) or unfavourable (red dotted line) prognosis, depending on EML4-ALK expression in CTCs (Log-rank test, P=0.017).
To verify the accuracy of our test, we performed FISH analysis (the “gold standard”) for EML4-ALK translocation in all the available biopsy specimens (11 out of 35 patients), in whom the FISH analysis confirmed the presence of EML4-ALK translocation.
Interestingly, not all the detected CTCs counted in the same sample were EML4-ALK-positive, further confirming that in PB CTCs are heterogeneous.
Notably, we could associate EML4-ALK-positive CTCs with shorter PFS (0.57 year) compared to NSCLC that did not express EML4-ALK in CTCs (0.94 year; Kaplan-Meier, Log-rank test, P=0.017) (Figure 5B). Moreover, we observed that after the first cycle of treatment and at radiological assessment, the number of EML4-ALK-positive CTCs was reduced or disappeared, whilst at progression the ELM4-ALK-positive CTCs reappeared, hence extending what we previously documented in three representative case reports (25,26) (see Supplementary files, Figure S3).
Discussion
In NSCLC, liquid biopsy is an attractive concept, since a simple blood test could inform about cancer, prognosis, and treatment effectiveness, potentially providing early evidence of drug resistance.
Using CS, previous studies detected only 35% CTC-positive patients in metastatic NSCLC (3,4), whilst the use of EpCAM-independent methods increased the rate of CTC detection (37,38).
Herein, we analysed a homogeneous cohort of advanced NSCLC; by SA we detected higher frequency of CTC-positive patients (53.6%), in comparison with previous reported rate ranging from 21 to 41% (3) for EpCAM-dependent assays. A more severe disease condition may explain this higher prevalence of CTC-positive patients in GR cohort by SA.
Furthermore, unlike the studies by Krebs and Allard (4,5), using SA we demonstrated that 1 CTC/7.5 mL of blood allows stratifying patients in two groups that significantly differ for prognosis (1.62 vs. 0.83 years, CTC-negative vs. CTC-positive, respectively; Kaplan-Meyer, Log-Rank test P=0.007). This data further confirms the more severe disease condition of GR cohort, since the EPAC-lung study, which comprised the GR cohort, recognized a poor prognostic value to 2 CTCs/7.5 mL PB (10).
In NSCLC, the CK pattern differs from other carcinomas (14). Hence, we repeated the CTC enumeration, in parallel, in a second blood draw collected at baseline from each patient, using our customized EA that recognizes an expanded CK profile, namely CK 4, 5, 6, 7, 8, 10, 13, 14, 18, 19, most of which are not included in the SA.
We found that EA allowed increasing the CTC-positive NSCLC up to 63.9% of patients, being the number of CTC-positive patients by EA significantly greater (115 out of 180) than CTC-positive patients by SA (103 out of 192; Chi-square =4.0179, with 1 degrees of freedom, P=0.04502).
Moreover, the findings that EA revealed a significant higher prevalence of never smoking women, which had at least 1 EML4-ALK-positive CTC/7.5 mL PB, (Fisher’s Exact test, P=0.00233) suggests the use of EA in never smoking women, to support clinicians in treatment decision.
However, to validate any novel assay for CTC enumeration (6), it is mandatory to demonstrate the biological significance of any reported increased sensitivity. Here, we show that, according to appropriate cut-off values, the EA was still able to stratify NSCLC patients in two groups, which differed for both PFS and OS, a criteria that is mandatory to implement a new CTC assay in clinical practice (6). In fact, since different CTC subpopulations might differ for metastatic potential (30), the mere comparison of metrics/blood unit between gold standard SA and novel assay EA, is not enough to support the superiority of the second one, simple based on higher prevalence of CTC-positive patients (6).
Anyway, what is most important to stress here is that by extending the CK examined, our EA discloses higher number of CTC-positive patients; hence, in principle, an increased number of patients could benefit of detecting “druggable” mutation in CTCs.
To test this hypothesis, we then established an integrated assay of EA to highlight the EML4-ALK fusion protein on CTCs, because of its potential utility in driving cancer treatment (25,26).
Indeed, to choose the appropriate first-line treatment, detection of EGFR mutation and ALK rearrangement are recommended as routine genetic profiling for non-squamous NSCLC or non-smoking populations. FISH, RT-PCR and Ventana IHC—with validated mAbs—are all currently accepted methods for the detection of ALK rearrangement in biopsies. Hence, characterizing CTCs could offer new perspectives for both the diagnosis and the monitoring of ALK-positive patients, eligible for ALK inhibitors.
To date, two groups demonstrated the feasibility to detect ALK rearrangement on CTCs, when enriched by filtrations (39,40). Our procedure differs from previous reports, and we think it seems to be more promising for clinical application. In fact, it permits to quantify directly both the CTC burden in peripheral blood and the fusion protein expression at single cell level, through a robust automated platform that allows the multiple measurements, required to monitor treatment effectiveness in individual patient.
Our EML4-ALK integrated assay proved to be highly sensitive, since we could confirm in all the available biopsy specimens (11 out of 35 patients), by FISH analysis, the presence of EML4-ALK translocation, as well in positive but not in negative controls during the assay’s development (Supplementary materials, Figure S1).
Although our frequency of EML4-ALK-positive samples is high—close to 30% vs. a frequency close to 5% reported in literature—it is similar to that reported in a selected group of non-squamous lung cancers occurred in never-smoking females, and tested with a highly performing IHC (41).
Furthermore, if it uses a robust method, it is conceivable that the heterogeneous expression of a given protein may be potentially more evaluable at single-cell level in peripheral blood. Indeed, it is intriguing to notice that, in our case, not all the CTCs detected in the same sample, expressed the EML4-ALK protein, reflecting tumour heterogeneity.
Using our integrated assay specific for EML4-ALK detection, we could not only detect EML4-ALK on CTCs but also relate them to a significant shorter PFS (0.57 vs. 0.94 years, in EML4-ALK CTC-positive patients vs. negative, respectively, P=0.017). Our success in establish, for the first time, a cut-off value of 1 EML4-ALK-positive cell for PFS depends both on the automation of our test, and on the use of an EpCAM-based technology. Indeed, it was recently reported that in NSCLC the CTC number increased when considering both EpCAMhigh and EpCAMlow CTCs, but only the first ones, and not the second ones strongly correlated with poor OS (31).
The main limit of our study is the incomplete record of some demographic characteristics of the GR cohort, so that some adjusted analyses were not significant anymore, since a lower sample size was available. Similarly, the lack in reporting disease progression determined the loss of follow-up in some cases, so that PFS and OS analyses differ for sample size.
However, our findings are of potential utility for clinicians, since EA identify higher number of CTC-positive patients, which might benefit from CTC monitoring.
It is essential and interesting to continue this deep characterization at successive time points, in order to evaluate the changing of mutational profile and the feasibility to investigate the evolution of disease in this compartment.
Indeed, by using our EML4-ALK-integrated assay, we have already provided the proof of concept that serial determination of EML4-ALK-positive CTCs was achievable in patients treated with ALK inhibitors and could anticipate the emergent drug resistance (25). Herein, the demonstration of the prognostic value of this test in CTCs offers a strong rationale to use our EML4-ALK-integrated assay in prospective trials, devoted to investigate ALK-specific treatments.
Finally, the common effort of scientific community in standardizing CTC characterization, in the near future will allow to apply routinely CTC assays in every lung cancer patient. Our results show that the information obtained from CTCs is able to suggest a prompt intervention, as a treatment change, before what established by the current guidelines.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors thank Christina Drace (IOV-IRCCS, Padua, Italy) for English editing.
Funding: This work was supported by grant from the Italian Ministry of Health, Proposal No: # GR-2010-2303193A, “Individualized treatments of patients with advanced NSCLC: potential application for CTCs molecular and phenotypical profiling” (PI: E.R.). This funding source had no role in the design of this study and will not have any role during its execution, analyses, interpretation of the data, or decision to submit results.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol [IOV-IRCCS study “Individualized treatment of patients with advanced NSCLC: potential application for circulating tumour cells (CTCs) molecular and phenotypical profiling”, NCT02407327] conforms to the provisions in accordance with the Helsinki Declaration as revised in 2013, and has been approved by IOV-IRCCS Ethical Committee (protocol # CE IOV 2012/52). PB samples were collected upon signed informed consent. Patients’ samples have been anonymized and the coding was known to researchers involved in the study, only to purposes of patients’ data traceability. Patients were awareness of the altruistic significance of their participation and in any case the study outcomes did not affect the next management of their treatment.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-855
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-855
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-855
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-855). Dr. ER reports grants from Italian Ministry of Health, during the conduct of the study. The other authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Morgensztern D, Ng SH, Gao F, et al. Trends in stage distribution for patients with non-small cell lung cancer: a National Cancer Database survey. J Thorac Oncol 2010;5:29-33. 10.1097/JTO.0b013e3181c5920c [DOI] [PubMed] [Google Scholar]
- 3.Hanssen A, Loges S, Pantel K, et al. Detection of Circulating Tumor Cells in Non-Small Cell Lung Cancer. Front Oncol 2015;5:207. 10.3389/fonc.2015.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- 5.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. 10.1200/JCO.2010.28.7045 [DOI] [PubMed] [Google Scholar]
- 6.Parkinson DR, Dracopoli N, Petty BG, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med 2012;10:138. 10.1186/1479-5876-10-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka F, Yoneda K, Kondo N, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res 2009;15:6980-6. 10.1158/1078-0432.CCR-09-1095 [DOI] [PubMed] [Google Scholar]
- 8.Ma XL, Xiao ZL, Liu L, et al. Meta-analysis of circulating tumor cells as a prognostic marker in lung cancer. Asian Pac J Cancer Prev 2012;13:1137-44. 10.7314/APJCP.2012.13.4.1137 [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Wang K, Xu J, et al. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PLoS One 2013;8:e78070. 10.1371/journal.pone.0078070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindsay CR, Blackhall FH, Carmel A, et al. EPAC-lung: pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur J Cancer 2019;117:60-8. 10.1016/j.ejca.2019.04.019 [DOI] [PubMed] [Google Scholar]
- 11.Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (Basel) 2014;6:153-65. 10.3390/cancers6010153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. 10.1158/1078-0432.CCR-11-3148 [DOI] [PubMed] [Google Scholar]
- 13.Pailler E, Faugeroux V, Oulhen M, et al. Routine clinical use of circulating tumor cells for diagnosis of mutations and chromosomal rearrangements in non-small cell lung cancer-ready for prime-time? Transl Lung Cancer Res 2017;6:444-53. 10.21037/tlcr.2017.07.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Cui T, Yang L, et al. The diagnostic value of cytokeratin 5/6, 14, 17, and 18 expression in human non-small cell lung cancer. Oncology 2011;80:333-40. 10.1159/000329098 [DOI] [PubMed] [Google Scholar]
- 15.Went PT, Lugli A, Meier S, et al. Frequent EpCam protein expression in human carcinomas. Hum Pathol 2004;35:122-8. 10.1016/j.humpath.2003.08.026 [DOI] [PubMed] [Google Scholar]
- 16.Fong D, Seeber A, Terracciano L, et al. Expression of EpCAM(MF) and EpCAM(MT) variants in human carcinomas. J Clin Pathol 2014;67:408-14. 10.1136/jclinpath-2013-201932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. 10.1097/JTO.0b013e31823c5c16 [DOI] [PubMed] [Google Scholar]
- 18.Kulasinghe A, Kapeleris J, Kimberley R, et al. The prognostic significance of circulating tumor cells in head and neck and non-small-cell lung cancer. Cancer Med 2018;7:5910-9. 10.1002/cam4.1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Kulasinghe A, Bogseth A, et al. Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel. Microsyst Nanoeng 2019;5:8. 10.1038/s41378-019-0045-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulasinghe A, Kapeleris J, Cooper C, et al. Phenotypic Characterization of Circulating Lung Cancer Cells for Clinically Actionable Targets. Cancers (Basel) 2019;11:380. 10.3390/cancers11030380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulasinghe A, Lim Y, Kapeleris J, et al. The Use of Three-Dimensional DNA Fluorescent In Situ Hybridization (3D DNA FISH) for the Detection of Anaplastic Lymphoma Kinase (ALK) in Non-Small Cell Lung Cancer (NSCLC) Circulating Tumor Cells. Cells 2020;9:1465. 10.3390/cells9061465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen S, Lo TW, Fouladdel S, et al. Simultaneous Single Cell Gene Expression and EGFR Mutation Analysis of Circulating Tumor Cells Reveals Distinct Phenotypes in NSCLC. Adv Biosyst 2020;4:e2000110. 10.1002/adbi.202000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofman V, Heeke S, Marquette CH, et al. Circulating Tumor Cell Detection in Lung Cancer: But to What End? Cancers (Basel) 2019;11:262. 10.3390/cancers11020262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pezzuto A, Manicone M, Scaini MC, et al. What information could the main actors of liquid biopsy provide? -a representative case of non-small cell lung cancer (NSCLC). J Thorac Dis 2018;10:E570-6. 10.21037/jtd.2018.06.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aieta M, Facchinetti A, De Faveri S, et al. Monitoring and Characterization of Circulating Tumor Cells (CTCs) in a Patient With EML4-ALK-Positive Non-Small Cell Lung Cancer (NSCLC). Clin Lung Cancer 2016;17:e173-7. 10.1016/j.cllc.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 26.Manicone M, Scaini MC, Rodriquenz MG, et al. Liquid biopsy for monitoring anaplastic lymphoma kinase inhibitors in non-small cell lung cancer: two cases compared. J Thorac Dis 2017;9:S1391-6. 10.21037/jtd.2017.08.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gennari A, Foca F, Zamarchi R, et al. Insulin-like growth factor-1 receptor (IGF-1R) expression on circulating tumor cells (CTCs) and metastatic breast cancer outcome: results from the TransMYME trial. Breast Cancer Res Treat 2020;181:61-8. 10.1007/s10549-020-05596-4 [DOI] [PubMed] [Google Scholar]
- 28.Pantano F, Rossi E, Iuliani M, et al. Dynamic changes of Receptor activator of nuclear factor-kappaB expression in Circulating Tumor Cells during Denosumab predict treatment effectiveness in Metastatic Breast Cancer. Sci Rep 2020;10:1288. 10.1038/s41598-020-58339-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franken A, Honisch E, Reinhardt F, et al. Detection of ESR1 Mutations in Single Circulating Tumor Cells on Estrogen Deprivation Therapy but Not in Primary Tumors from Metastatic Luminal Breast Cancer Patients. J Mol Diagn 2020;22:111-21. 10.1016/j.jmoldx.2019.09.004 [DOI] [PubMed] [Google Scholar]
- 30.Allard WJ, Terstappen LW. CCR 20th Anniversary Commentary: Paving the Way for Circulating Tumor Cells. Clin Cancer Res 2015;21:2883-5. 10.1158/1078-0432.CCR-14-2559 [DOI] [PubMed] [Google Scholar]
- 31.de Wit S, Dalum Gv, Lenferink ATM, et al. The detection of EpCAM+ and EpCAM– circulating tumor cells. Scientific Reports 2015;5:12270. 10.1038/srep12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi E, Basso U, Celadin R, et al. M30 neoepitope expression in epithelial cancer: quantification of apoptosis in circulating tumor cells by CellSearch analysis. Clin Cancer Res 2010;16:5233-43. 10.1158/1078-0432.CCR-10-1449 [DOI] [PubMed] [Google Scholar]
- 33.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 34.Rowand JL, Martin G, Doyle GV, et al. Endothelial cells in peripheral blood of healthy subjects and patients with metastatic carcinomas. Cytometry A 2007;71:105-13. 10.1002/cyto.a.20364 [DOI] [PubMed] [Google Scholar]
- 35.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 36.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. 10.1056/NEJMoa1006448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang T, Jia CP, Jun Y, et al. Highly sensitive enumeration of circulating tumor cells in lung cancer patients using a size-based filtration microfluidic chip. Biosens Bioelectron 2014;51:213-8. 10.1016/j.bios.2013.07.044 [DOI] [PubMed] [Google Scholar]
- 38.Hofman V, Long E, Ilie M, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology 2012;23:30-8. 10.1111/j.1365-2303.2010.00835.x [DOI] [PubMed] [Google Scholar]
- 39.Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. 10.1093/annonc/mds137 [DOI] [PubMed] [Google Scholar]
- 40.Faugeroux V, Pailler E, Auger N, et al. Clinical Utility of Circulating Tumor Cells in ALK-Positive Non-Small-Cell Lung Cancer. Front Oncol 2014;4:281. 10.3389/fonc.2014.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han XH, Zhang NN, Ma L, et al. Immunohistochemistry reliably detects ALK rearrangements in patients with advanced non-small-cell lung cancer. Virchows Arch 2013;463:583-91. 10.1007/s00428-013-1472-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as