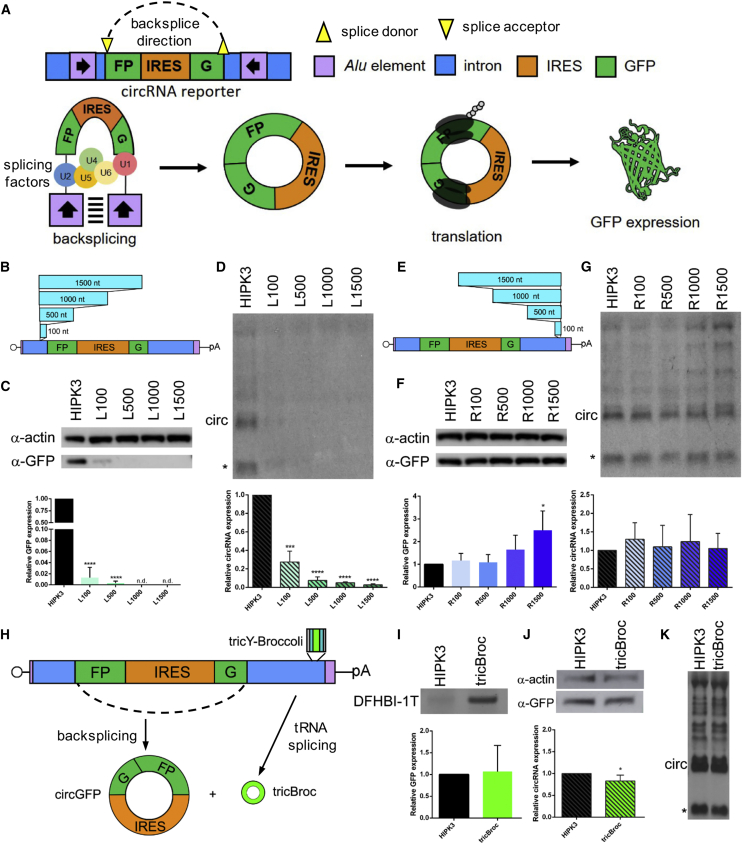
Figure 1.
Distance requirements between Alu element and splice site differ in upstream and downstream introns
(A) A reporter was constructed by extracting portions of introns from the HIPK3 gene and placing them around a split GFP reporter exon. Inverted Alu repeats in the introns interact, allowing for backsplicing to occur, forming a circRNA. The presence of an IRES sequence drives translation, leading to GFP protein expression. See Figure S1 for intron sequences. (B) Sequence ranging from 100 nt to 1,500 nt was inserted into the left HIPK3 intron at the indicated position. See also Figure S2. (C and D) Constructs were transfected into HEK293 cells and expression assayed at 4 days post-transfection by (C) western blot analysis, with actin as a loading control (quantification below), and (D) northern blot analysis, probing for GFP sequences (quantification below). (E) Sequence ranging from 100 nt to 1,500 nt was inserted into the right HIPK3 intron at the indicated position. (F and G) Constructs were transfected into HEK293 cells and expression assayed at 4 days post-transfection by (F) western blot analysis, with actin as a loading control (quantification below), and (G) northern blot analysis, probing for GFP sequences (quantification below). (H) Sequences driving formation of a tricRNA containing Broccoli (tricY-Broccoli) were inserted into the same location in the right HIPK3 intron. (I) tricY-Broccoli expression was verified by gel electrophoresis followed by DFHBI-1T staining. (J and K) circRNA formation was assayed by (J) western blot analysis, with actin as a loading control (quantification below left), and (K) northern blot analysis, probing for GFP sequences (quantification left). On northern blots, the asterisk refers to an additional circular band. Western and northern blots were quantified as detailed in Materials and methods and graphed relative to the unchanged HIPK3 intron construct. Student’s t test was performed to test for statistical significance. ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005; ∗∗∗∗p < 0.00005.