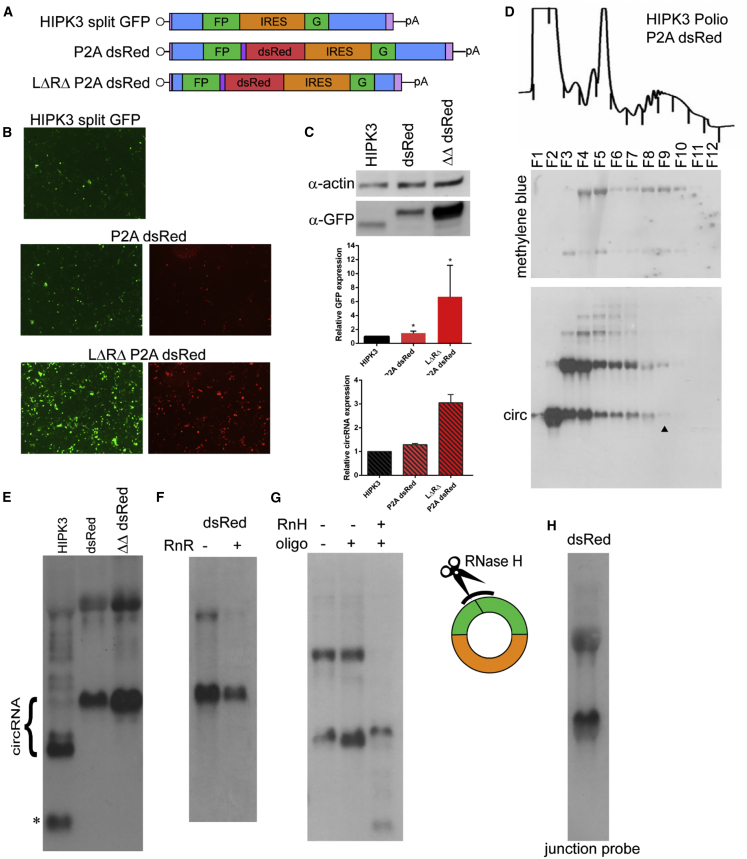
Figure 4.
circRNA size can be increased without loss of expression
(A) A self-cleaving P2A peptide followed by the dsRed ORF was added to the exon at the end of the GFP fragment either in the original HIPK3 construct or in the LΔRΔ intron pair. (B and C) Constructs were transfected into HEK293 cells and expression assayed at 4 days post-transfection by (B) GFP and dsRed fluorescence and (C) western blot analysis, with actin as a loading control (quantification below). (D) To assay translation efficiency, the construct was transfected into HEK293 cells and harvested in cycloheximide followed by a sucrose gradient and fractionation. Top: OD trace of the gradient, with fractions marked by lines. Middle: RNA was extracted from gradients, separated by gel electrophoresis, transferred to a membrane, and stained with methylene blue to visualize the ribosomal RNA. Bottom: the same membrane was probed for GFP sequences. The arrowhead marks the last fraction in which the circRNA is detected. (E) RNA from transfected cells was analyzed by northern blot analysis, probing for GFP sequences (quantification above right). (F) RNA was treated with RNase R, then analyzed by northern blot, probing for GFP sequences. (G) RNase H (RnH) digestion was performed with an oligonucleotide targeting the backsplice junction. Samples were analyzed by northern blot and probed against GFP sequences. (H) RNA was analyzed by northern blot and probed with an oligonucleotide spanning the backsplice junction. On northern blots, the asterisk refers to an additional circular band. Western and northern blots were quantified as detailed in Materials and methods and graphed relative to the unchanged HIPK3 intron construct. Student’s t test was performed to test for statistical significance. ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005; ∗∗∗∗p < 0.00005.