Abstract
Long intergenic noncoding RNAs (lincRNAs) are strongly associated with several kinds of cancer, including gastric cancer. Here, we found significantly decreased lincRNA-01317 levels in cancer tissue compared with paracancer tissue of patients with gastric cancer, and lincRNA-01317 expression levels positively correlated with clinical survival rate. Furthermore, using a gastric cancer cell line and a xenograft mouse model, we found that transfection of a gastric cancer cell line with lincRNA-01317 significantly inhibited the proliferation, migration, and invasion of gastric cancer cells. Finally, we demonstrated that lincRNA-01317 may target KCNQ1, as KCNQ1 was downregulated after transfection of cells with lincRNA-01317. This study aimed to assess lincRNA-01317 as a potential therapeutic target to treat cancers.
Keywords: LincRNA01317, gastric cancer
Introduction
Gastric cancer is the third leading cause of cancer-related deaths and also one of the most prevalent cancers in Eastern Asia [1]. Patients with gastric cancer often display no noticeable symptoms in the early stage; thus, when diagnosed as distant metastasis, gastric cancer is basically incurable at this stage. Targeted chemotherapy significantly improves the gastric cancer patients’ survival rate [2]. Nevertheless, severe adverse reactions and complications caused by chemotherapy add to the clinical challenge. Hence, the treatment for the gastric cancer patients requires the invention of new drugs and new combinations of treatments.
Long intergenic noncoding RNAs (lincRNAs) constitute a group of RNAs >200 nucleotides length with limited protein-coding ability [3]. Reportedly, lincRNAs play a vital role in cell biological function, such as development, differentiation, proliferation, metabolism, and invasion [4-7]. Moreover, they are closely associated with gastric cancer occurrence, invasion, and metastasis [8,9]. The lincRNAs, particularly lincRNA-01317, are novel and non-annotated [10], and of great research value.
Our study demonstrated that lincRNA-01317 is significantly downregulated in cancerous tissue compared with paracancerous tissue. After transfection of gastric cancer cell lines with lincRNA-01317, migration, proliferation, and invasion of the cells were compromised, indicating that lincRNA-01317 play an important role in the pathogenesis of gastric cancer and is a potential therapeutic target.
Materials and methods
Human samples
Cancer and paracancer (>5 cm from the edge of the tumor) tissue samples from nine patients who underwent radical gastrectomy for gastric cancer at the Shanghai General Hospital of the Nanjing Medical University from September 2018 to November 2018. Patients (mean age: 52.9 years; range: 26-70 years) underwent neither chemotherapy nor radiotherapy before the surgery. Table 1 summarizes the characteristics of the patients with gastric cancer enrolled in this study. The gastric cancer tissue microarray was derived from 55 patients with gastric cancer who underwent surgical resection between June 2010 and May 2011, in Shanghai General Hospital and all patients were followed until May 2019. The detailed clinical information for the patients is listed in Table 2. The experimental protocol was approved by the Ethics Committee of the Shanghai General Hospital. Before enrollment in the study, each patient signed an informed consent.
Table 1.
Clinical and demographic characteristics of the patients for the study of lncRNAs in tumor tissues and adjacent tissues
| Patient no. | Age (years) | Gender | Stage | Grade |
|---|---|---|---|---|
| 1 | 47 | F | T3N1M0 | I |
| 2 | 46 | M | T4N0M0 | I |
| 3 | 50 | M | T4N2M0 | II |
| 4 | 68 | F | T4N2M0 | II |
| 5 | 70 | F | T1N0M0 | I |
| 6 | 56 | M | T2N0M0 | II |
| 7 | 51 | M | T3N0M0 | II |
| 8 | 62 | M | T4N1M0 | II |
| 9 | 26 | F | T4N0M0 | II |
Table 2.
Clinicopathologic features in 55 patients with gastric cancer
| Variable | No. of patients | lincRNA-01317 density | P | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age, y | ||||
| < 65 | 24 | 12 | 12 | 1.000 |
| ≥ 65 | 31 | 15 | 16 | |
| Gender | ||||
| Male | 28 | 15 | 13 | 0.952 |
| Female | 27 | 14 | 13 | |
| Differentiation | ||||
| Well/moderate | 25 | 10 | 15 | 0.745 |
| Poor | 30 | 20 | 10 | |
| Depth of invasion | ||||
| T1-T2 | 5 | 2 | 3 | 0.003 |
| T3-T4 | 50 | 40 | 10 | |
| Lymph node metastasis | ||||
| Yes | 51 | 40 | 11 | 0.0026 |
| No | 4 | 3 | 1 | |
| Tumor stage | ||||
| I-II | 10 | 8 | 2 | 0.0038 |
| III-IV | 45 | 40 | 5 | |
Mice
Four-week old female nude mice were obtained from the Model Animal Research Center of Nanjing Universityand housed in the animal experimental center of Shanghai General Hospital. The experimental procedures were in accordance with the Animal Care and Use Committee at Nanjing Medical University.
Cell lines and culture conditions
Human gastric cancer cell lines (AGS and HGC-27) were obtained from the Type Culture Collection of the Chinese Academy of Science (Shanghai, China). The cell lines were maintained in F12 or RPMI 1640 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin in a humidified incubators (5% CO2) at 37°C.
Public database
TCGAwebsite (https://cancergenome.nih.gov/) was used to search publicly available datasets for recent studies of gastric cancer. The distribution data of the most frequently mutated lincRNAs were obtained from male and female patients with whole human species.
RT-qPCR
Total RNA was extracted using the RNeasy Micro Kit (Qiagen, Hilden, Germany) and reversed transcribed into cDNA using the RT Reagent Kit (TaKaRa, Tokyo, Japan). RT-qPCR amplification was performed in a 20-mL reaction volume containing cDNA, primers, and SYBR Green I Supermix (TaKaRa) using an ABI 7500 Thermocycler (Applied Biosystems, Foster City, CA, USA). Supplementary Table 1 lists the primers for the target genes.
FISH
Unstained 5-μm sections or tissue paraffin chip was used for FISH analysis. LincRNA-01317 and lincRNA-00886 were detected using the single-color probe kit (Abbott Laboratories, Des Plaines, IL, USA). On the first day, the slides were pretreated and hybridized using the VP2000 (SciGene, Sunnyvale, CA, USA), per the manufacturer’s instructions. On the following day, the slides were washed using Little Dipper (Abbott Laboratories). The washed slides were counterstained with DAPI (Vector Laboratories, Burlingame, CA, USA) and cover slips were applied using Vectashield mounting medium. Furthermore, the slides were stored at -80°C if not scored immediately.
Lentiviral vector production, titration, and transduction
The full-length lincRNA-01317 sequence is the following: TGCCACCACGTAAGAAGTGCCTTTTGCCTCCCACCATTATTCTGAGGCCTCCTCAGCCATGTGGAACTGATGACACATCTAGAAGACCCTCACCTTATGCTTATGCTGGCCCCTTGATCTTGGCCTCCAGAAATGTATAATGAAGAATCTTGAGGTCCTCCTGGATTACCAAAGTGGGCCCTAAATCCAGTGGCAGAAGACACAGACACAGAGAGGAGACCAGGTGAAGACAAAAGAAGAGGCTGGAGTGATGCAACCATCAGAGAGTGAAAATACTCAGTTACCTCCTGGTTTATGAGGCAATGAAGTATTCCAGCACATAAATATTGACGCTGAGACCCATGGGATGTAGGAGTGCTGAGCACATTTGCAAGACATAAAGACCGAAGAGGTGAATCACTTCAAAGAGAAGGACCAGATTCTCATGGACCCCACTGCCATGCTGGCCAGACATCTTGCTGCCATTGTTTTCCTGTTATTTACTCCCAAACAGATGGAGTGTCGCTCTGTCACCCAGGCTGGAGTGCAGTGGCACGATCTCGGCTTACTGCAAGATCTACCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACTACAGGCGTCCGCCACCACGCCCGGCTAATTTTTTGTTTTTTTTAGTATAGACGGGGTTTCACCGTGTTAGCCAGGATGGTCTCAATCTCCTGACCTCGTGACCCGCCCGCCTTGGCCTCCCAAAGTGAAGTCTCTTTTCAAAATCTTAATGCTTACCTGTGTTTCTTT. The sequence was synthesized and inserted into lentiviral vector pGCSIL-GFP with T4 DNA ligase. Competent Escherichia coli DH5α cells were transformed with the ligated vector. 293T cells were co-transfected with pGC-LV, pHelper 1.0, and pHelper 2.0 plasmids to produce the lentivirus. A549 cells were infected with lentivirus in medium containing polybrene (5 μg/mL). The positively infected cells were sorted by FACS to be further cultured. The level of lincRNA-01317 was detected by RT-qPCR [11].
Cell viability assays
Cells of control and experimental groups was seeded in each well of the plate to compare cell growth kinetics using the CCK-8 (Dojindo, Kumamoto, Japan) [12]. After incubating the plate for 24-96 h, measure the absorbance at 450 nm using a microplate reader (Thermo Scientific, Waltham, MA, USA), and the absorbance values were normalized to the values of the cells at 0 h.
Plate colony-formation assay
The 6-well plates were inoculated with 1000 cells per well and cultured for 7 days. After being fixed with paraformaldehyde, the colonies were stained with 5% crystal violet for >1 hour. The colonies were photographed and counted using a dissecting microscope. Colony-counting software was used to scan the images of each plate [13].
Cell migration and invasion assay
Cell migratory capacity was assessed using the 8 µm pore size 24-well transwell system (Corning, Corning, NY, USA). In this study, 3 × 105 cells were seeded in the upper chamber and allowed to migrate for 24 h. Cells in the chamber were fixed with methanol and stained with 0.1% crystal violet (C8470, Amresco, Dallas, TX, USA), and photographs were taken [13]. For the invasion assay, 2 × 104 cells were inoculated in serum-free medium in the upper chamber coated with Matrigel matrix (BD Biosciences, USA). After 24 h, the upper chambers were fixed and stained as described before. The cells that had migrated to the reverse side of the upper chambers were photographed [14]. Five fields were selected randomly to count migrated or invaded cells.
Cell wound healing
For the wound-healing experiment, the cells were first cultured to confluence in 6-well plates. The culture was then scratched in the center of the well with a 200-μL microtubule tip. The cells were washed with PBS and incubated with serum-free medium. After 24 h, representative images were captured. The width of the healed area was quantified and compared with the baseline values. All experiments were independently repeated in triplicate [14].
Xenograft model of human colon cancer
AGS or HGC-27 human colon cancer cells (5 × 106 cells in 100 mL phosphate-buffered saline [PBS] per mouse) were injected intraperitoneally into the flanks of mice. The tumor volume was calculated using the formula (length × width2 × 0.5) every three days. At day 10, the mice were euthanized with pentobarbital sodium (200 mg/kg), and tumors were excised for further examination.
Prediction of lincRNA-01317 binding
The website http://rtools.cbrc.jp/ was used to predict the binding target-gene or RNA-of lincRNA-01317. The results were verified by RT-qPCR.
Western blot analysis
Cell were harvested and denatured. Proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred ontopolyvinylidene difluoride membranes. Membranes were blocked and incubated overnight at 4°C with anti-KCNQ1 antibody or anti-YIPF antibody (EP1674Y; Abcam, Cambridge, UK). The target protein was detected by an enhanced chemiluminescence western blot detection system (Thermo Fisher Scientific).
Statistical analysis
Results obtained for the two groups in the study were compared using Student’s t-test. Statistical analysis was performed using GraphPad software (GraphPad, San Diego, CA, USA). P < 0.05 was considered statistically significant.
Results
LincRNA-01317 was frequently downregulated in gastric cancer tissue
Globally, the survival rate for gastric cancer is quite low. According to the TCGA tumor database, the five-year survival rate for gastric cancer is 40%, whereas the ten-year survival rate is only 20%. The TCGA database was used to detect lincRNAs that are most prone to mutation in gastric cancer, and their expression in cancerous and paracancerous tissue was verified. The 20 genes that were most susceptible to mutation were selected for the study (Supplementary Table 2). RT-qPCR analysis revealed that two lincRNAs (lincRNA-01317 and lincRNA-00886) were downregulated in the cancerous tissues obtained from the patients compared with the paracancerous tissue samples obtained from the same patients (Figure 1A and 1B). The significance of the observed differences in their expression levels was assessed using Student’s t-test, based on the presence or absence of a normal distribution in the data, and revealed significant differences in the expression of lincRNA-01317 and lincRNA-00886 between the groups (P = 0.0315 and P = 0.0464, respectively); however, no significant difference was observed in other mutation-prone lincRNAs (Supplementary Figure 1). FISH was performed in three paired biopsy specimens of patients with gastric cancer to validate the expression levels of lincRNA-01317 and lincRNA-00886 in gastric cancer. Figure 1C and 1D show that lincRNA-01317 expression was markedly increased, along with a higher fluorescence value, and lincRNA-00886 expression was relatively unchanged in the paracancerous tissue compared with the corresponding cancerous tissue. Thus, we focused on the role of lincRNA-01317 in gastric cancer.
Figure 1.
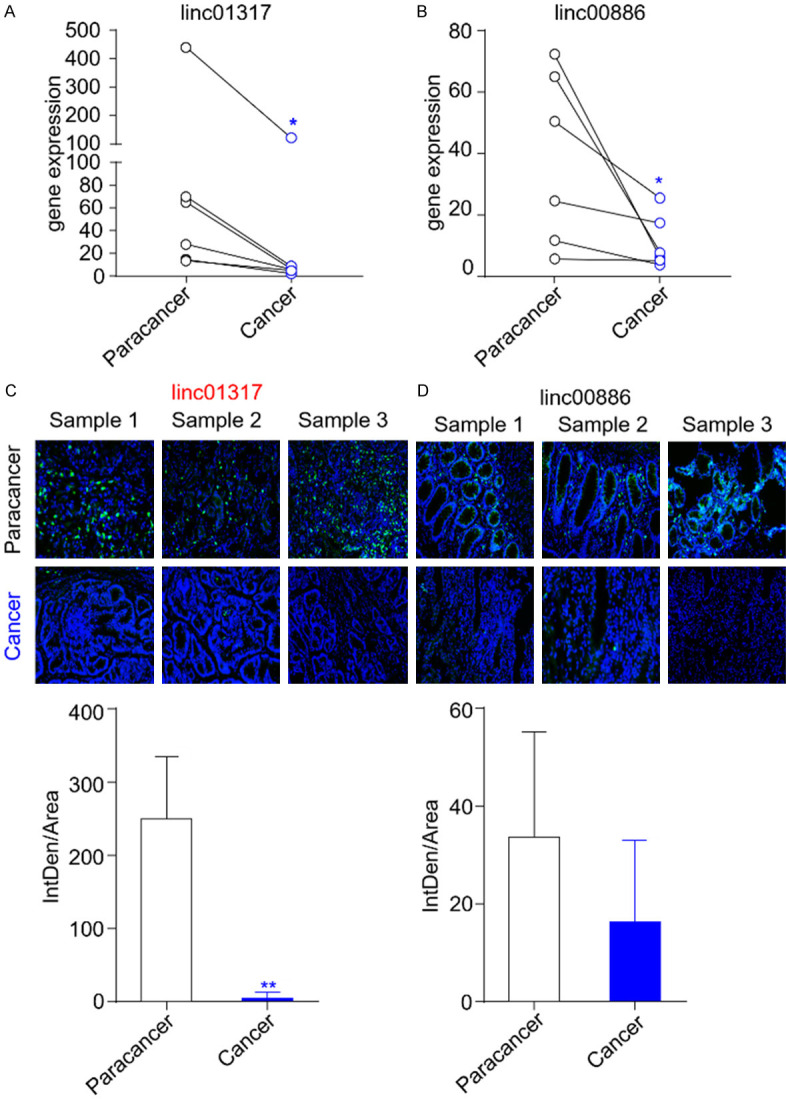
LincRNA-1317 is downregulated in gastric cancer tissue. Levels of lincRNA-01317 (A) and lincRNA-00886 (B) in six pairs of gastric cancer tissue and corresponding paracancer tissue were detected using RT-qPCR. The RT-qPCR results are shown as the mean ± standard error of the mean (SEM). (C) In situ hybridization of lincRNA-01317 in paracancer tissue and gastric cancer tissue was performed. Green: lincRNA-01317; blue: nuclei counterstained with 40,6-diamidino-2-phenylindole (DAPI). (D) In situ hybridization of lincRNA-00886 in paracancer and gastric cancer tissues was performed. Green: lincRNA-00886; blue: DAPI. The statistical results are shown as the mean ± SEM (*P < 0.05 and **P < 0.01).
Downregulation of lincRNA-01317 positively correlates with poor prognosis in gastric cancer patients
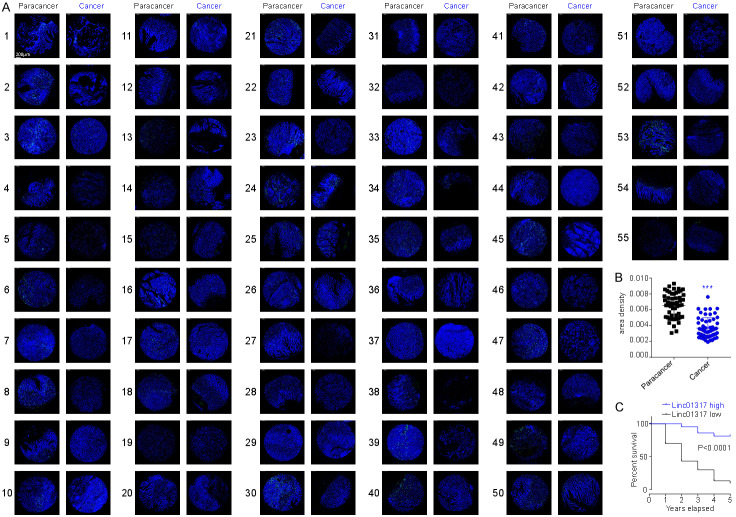
To explore the clinical significance of lincRNA-01317 in gastric cancer, 55 cancer specimens were detected by FISH, and the correlation between lincRNA level and the clinical characteristics was analyzed. The results showed that lincRNA-01317 was downregulated in cancer tissue (Figure 2A and 2B). Moreover, patients with low levels of lincRNA-01317 expression showed lower 5-year disease-specific survival rate than those with high lincRNA-01317 expression (Figure 2C). Furthermore, levels of lincRNA-01317 were negatively correlated with the tumor stage and lymph node metastasis (Table 2), suggesting that downregulation of lincRNA-01317 is an independent indicator of prognosis in gastric cancer.
Figure 2.
LincRNA-01317 is downregulated in cancer tissue from patients with gastric cancer and is an indicator of poor prognosis. (A) In situ hybridization of lincRNA-01317 in paracancer and cancer tissue from 55 advanced-stage gastric cancer patients. (B) The results of statistical analysis of the data in (A) are shown as the mean ± SEM (***P < 0.001). (C) Low expression level of lincRNA-01317 significantly (log rank, P < 0.0001) correlates with poor progression-free survival in this patient cohort.
Overexpression of lincRNA-01317 suppressed proliferation, migration, and invasion of a gastric cancer cell line in vitro
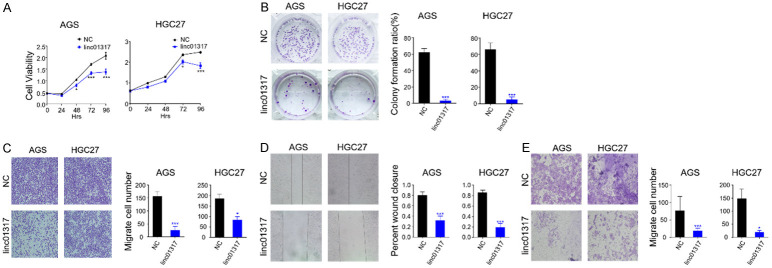
Given the low expression level of lincRNA-01317 in gastric cancer tissue, we further evaluated the biological role played by this lincRNA in gastric cancer cell lines. We stably transfected AGS and HGC-27 cells with the synthetic lincRNA-01317 sequence (Supplementary Figure 2). For controls, we transfected cancer cells with negative control lincRNA (lincRNA-NC) that did not specifically target any human gene products. After 48 h, the growth of lincrna-01317 transfected cells in both cell lines was slower than that in the NC group. As in the CCK-8 proliferation assay, the proliferation capacity of AGS and HGC-27 cells decreased in the presence of lincRNA-01317 (Figure 3A). In addition, overexpression of lincRNA-01317 inhibited the rate of colony formation (Figure 3B). To further understand the role of lincRNA-01317 in gastric cancer, transwell migration (Figure 3C), wound healing (Figure 3D), and invasion (Figure 3E) assays were performed. The data show that overexpression of lincRNA-01317 inhibits the migration and invasion of AGS and HGC-27 cells gastric cancer cells.
Figure 3.
LincRNA-01317 inhibits AGS and HCG27 cell proliferation, migration, and invasion. (A) Stable lincRNA01317-expressing AGS and HCG27 cells were established and linc-NC-expressing cells were control groups. Cell viability and proliferation were evaluated by the CCK-8 assay at 24, 48, 72, and 96 h. Results of statistical analysis are shown as the mean ± SEM (*P < 0.05 and ***P < 0.001). (B) The proliferative ability of lincRNA-01317-expressing AGS and HCG27 cells and NC control cells was assessed via a plate colony-formation assay. The results of statistical analysis are shown as the mean ± SEM (***P < 0.001). (C) LincRNA-01317-expressing AGS and HCG27 cells could migrate on transwell inserts for 24 h. The cells that had migrated into the lower chambers through the filter were quantitated by gentian violet staining and expressed as the total number of cells in the lower wells. Cell mobility and invasion were assessed by wound-healing assay (D) and transwell migration through Matrigel. (E) The results of statistical analysis are shown as the mean ± SEM (*P < 0.05 and ***P < 0.001).
LincRNA-01317 inhibits tumor growth in a mouse xenograft model of gastric cancer
Given the suppression of cancer cell migration and invasion by lincRNA-01317 in vitro, we further assessed whether increased levels of lincRNA-01317 would inhibit tumor formation by gastric cancer cells in vivo. LincRNA-01317-overexpressing AGS and HGC-27 cells were subcutaneously implanted into either the left or right posterior flank, respectively, of the same nude mouse (5 × 106 cells per injection site) (Figure 4A). Six days after implantation, the cells transfected with linc-NC formed tumors, while the cells transfected with lincRNA-01317 failed to grow or grew slowly (Figure 4A), exhibited a marked reduction in tumor size and tumor weight (Figure 4B and 4C) compared with the control group. These data indicated that elevated lincRNA-01317 levels in gastric cancer cells markedly reduced their ability to form tumors.
Figure 4.
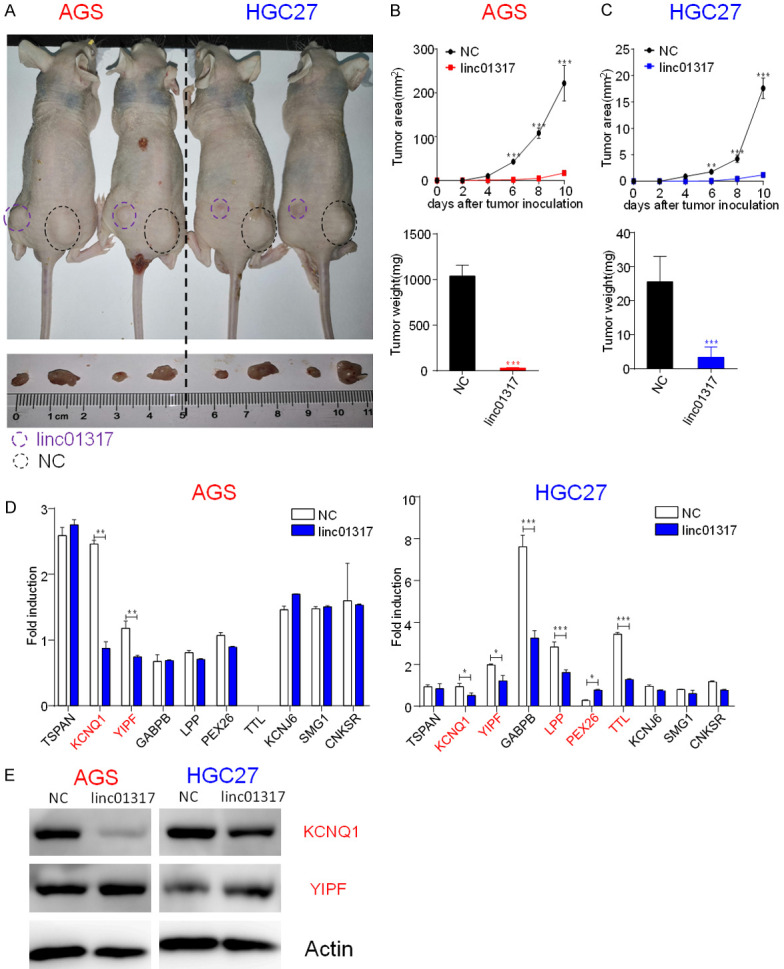
LincRNA-01317 inhibited AGS and HCG27 cell tumor growth in vivo. LincRNA- and linc-NC-transfected AGS and HCG27 cells (5 × 106) were injected subcutaneously into the left or right posterior flank, respectively, of the same nude mouse (as indicated). A. Photographs were taken 10 days after tumor-cell implantation. B. Tumor sizes were measure with a vernier caliperare and were shown as mean ± SEM. C. Tumor weight is also shown as mean ± SEM. Four mice were used in each experiment and repeated twice, for a total of eight or more mice. D. Candidate target gene expression measured by RT-qPCR in lincRNA-01317 and linc-NC-transfected AGS and HCG27 cells. Data are representative of two independent experiments (*P < 0.05 and ***P < 0.001). Western analysis showed that levels of total and cleaved caspase-3 were significantly downregulated in both lincRNA-01317-expressing cell lines. β-actin served as control. D. KCNQ1 and YIPF expression levels were measured by western analysis in lincRNA-01317- and linc-NC-transfected AGS and HCG27 cells. KCNQ1 was downregulated in both lincRNA-01317-expressing cell lines. E. Western analysis of levels of KCNQ1 and YIPF. β-actin was used as the control.
KCNQ1 emerged as a novel target of lincRNA-01317
Based on the literature [15], we hypothesized that lincRNA-01317 may inhibit the malignant phenotype of gastric cancer cells by regulating genes that control cell proliferation, migration, or invasion. Therefore, we decided to use rtools to find the target RNAs of lincRNA-01317. We found that a variety of genes had binding sites (Supplementary Table 3), and we selected 10 RNAs with the highest scores for RT-qPCR verification, among which KCNQ1 and YIPF showed changes in both cell lines and showed decreased expression in lincRNA-01317-overexpressing cell lines (Figure 4D). We then measured the expression of these two genes in the gastric cancer and paracancer tissues, and showed that KCNQ1 expression in the tumor decreased (Supplementary Figure 3A), which was positively correlated with the expression of lincRNA-01317 (Supplementary Figure 3B). Western analysis also confirmed that KCNQ1 levels decreased significantly in the tumor cells (Figure 4E). Taken together, these results suggested that lincRNA-01317 reduces the expression of KCNQ1 via the interactions.
Discussion
Increasing evidence has indicated that lincRNAs are critical regulators of cancer-related processes. LincRNA expression shows close correlation with various cancers, and they are function as either tumor suppressor genes or oncogenic genes [16]. During tumor growth and progression, overexpressed or downregulated lincRNAs may potentially target tumor suppressor genes and/or oncogenic genes [17-20]. We first used the TCGA database to identify nearly 200 mutation-prone lincRNAs, and GO analysis revealed that most were new lincRNAs with unannotated functions. The lincRNAs that were most susceptible to mutation and had sequences available were selected. Furthermore, six pairs of tissues were collected from patients with gastric cancer, and RT-qPCR was performed, followed by a paired t-test. Two lincRNAs, lincRNA-01317 and lincRNA-00886, were determined to be expressed at a high level in the paracancerous tissues, but at a relatively low expression in the cancerous tissues. Thus, three additional biopsy specimens of patients with gastric cancer were collected and FISH was performed to validate the results. It was confirmed that lincRNA-01317 was expressed at a high level in the paracancerous tissue and at a markedly low level in the cancerous tissues. Furthermore, lincRNA-00886 was expressed at a negligible level in the paracancerous tissues and not at all in the cancerous tissue. Therefore, this suggests the involvement of lincRNA-01317 in the development and progression of gastric cancer and possible applications in diagnostics or targeted therapy. In gastric tissues, Yan et al. showed that lincRNA-00470 promotes tumors growth [21] and linc-00629 inhibits tumor growth [22]. We observed a significant inhibition of cell growth, migration, and invasion with overexpression of lincRNA-01317, suggesting that lincRNA-01317 function as a tumor suppressor in gastric cell lines. Furthermore, lincRNA-01317 expression in tissue samples from 55 gastric cancer patients and the survival rates of the patients showed that lincRNA-01317 was closely correlated with the progression of gastric cancer and could be used as a new biological marker and target.
The general function of lincRNAs is to modulate their targets by direct or indirect binding of RNAs [17,18]. As these are new lincRNAs with unannotated functions, they are not described in the existing literature. Based on the bioinformatics analysis, we predicted several lincRNA-01317 targets. Our results suggested that KCNQ1, which is a tumor suppressor gene [23], might be a novel target of lincRNA-01317 in gastric cancer cells. Overexpression of lincRNA-01317 significantly down regulated KCNQ1 RNA level. In conclusion, this study may serve as a basis for future studies regarding biological characteristics and the applications of lincRNA-01317.
Acknowledgements
This work was supported by the Science and Technology Committee of Songjiang District, Shanghai (19SJKJGG140).
Disclosure of conflict of interest
None.
Abbreviations
- lincRNAs
Long intergenic noncoding RNAs
- TCGA
The Cancer Genome Atlas
- RT-qPCR
Quantitative reverse transcription-polymerase chain reaction
- FISH
fluorescence in situ hybridization
- CCK-8
cell counting Kit-8
Supporting Information
References
- 1.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Chen X, Zhang X, Wang L, Cao P, Rajamanickam V, Wu C, Zhou H, Cai Y, Liang G, Wang Y. Curcuminoid B63 induces ROS-mediated paraptosis-like cell death by targeting TrxR1 in gastric cells. Redox Biol. 2018;21:101061. doi: 10.1016/j.redox.2018.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan L, Wu X, Liu Y, Xian W. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J Cell Biochem. 2018 doi: 10.1002/jcb.27999. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Qi S, Duan Z, Liao H, Yang B, Wang W, Tan J, Li Q, Xia X. Long non-coding RNA LOC554202 modulates chordoma cell proliferation and invasion by recruiting EZH2 and regulating miR-31 expression. Cell Prolif. 2017;50:e12388. doi: 10.1111/cpr.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang XY, Zhang LX, Tian CJ, Tang XY, Zhao LM, Guo YL, Cheng DJ, Chen XL, Ma LJ, Chen ZC. LncRNAs BCYRN1 promoted the proliferation and migration of rat airway smooth muscle cells in asthma via upregulating the expression of transient receptor potential 1. Am J Transl Res. 2016;8:3409–3418. [PMC free article] [PubMed] [Google Scholar]
- 7.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanelli GN, Gasparini P, Coati I, Cui R, Pakula H, Chowdhury B, Valeri N, Loupakis F, Kupcinskas J, Cappellesso R, Fassan M. LONG-NONCODING RNAs in gastroesophageal cancers. Noncoding RNA Res. 2018;3:195–212. doi: 10.1016/j.ncrna.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li PF, Chen SC, Xia T, Jiang XM, Shao YF, Xiao BX, Guo JM. Non-coding RNAs and gastric cancer. World J Gastroenterol. 2014;20:5411–5419. doi: 10.3748/wjg.v20.i18.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10:103. doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Chu Y. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci. 2011;7:805–814. doi: 10.7150/ijbs.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soucheray M, Capelletti M, Pulido I, Kuang Y, Paweletz CP, Becker JH, Kikuchi E, Xu C, Patel TB, Al-Shahrour F, Carretero J, Wong KK, Janne PA, Shapiro GI, Shimamura T. Intratumoral heterogeneity in EGFR-mutant NSCLC results in divergent resistance mechanisms in response to EGFR tyrosine kinase inhibition. Cancer Res. 2015;75:4372–4383. doi: 10.1158/0008-5472.CAN-15-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, He J, Yang Z, Ge S, Zhang H, Zhong Q, Fan X. ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy. 2020;16:1186–1199. doi: 10.1080/15548627.2019.1659614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, Rong Z, Luo Z, Yu Z, Zhang J, Qiu Z, Huang C. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer. 2019;18:126. doi: 10.1186/s12943-019-1054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17:135–143. doi: 10.1007/s10142-016-0524-x. [DOI] [PubMed] [Google Scholar]
- 16.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Lu B, Ren S, Wu F, Wang X, Yan C, Wang Z. Long noncoding RNA LINC01116 contributes to gefitinib resistance in non-small cell lung cancer through regulating IFI44. Mol Ther Nucleic Acids. 2019;19:218–227. doi: 10.1016/j.omtn.2019.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Zheng J, Liu L. The long noncoding RNA PCGEM1 promotes cell proliferation, migration and invasion via targeting the miR-182/FBXW11 axis in cervical cancer. Cancer Cell Int. 2019;19:304. doi: 10.1186/s12935-019-1030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HC, Kang D, Han N, Lee Y, Hwang HJ, Lee SB, You JS, Min BS, Park HJ, Ko YG, Gorospe M, Lee JS. A novel long noncoding RNA Linc-ASEN represses cellular senescence through multileveled reduction of p21 expression. Cell Death Differ. 2020;27:1844–1861. doi: 10.1038/s41418-019-0467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, Ma A, Jin Y, Pan G, Wang C. LncRNA SNHG16 induced by TFAP2A modulates glycolysis and proliferation of endometrial carcinoma through miR-490-3p/HK2 axis. Am J Transl Res. 2019;11:7137–7145. [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W, Huang Z. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521:887–893. doi: 10.1016/j.bbrc.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Wang L, He F, Li B, Han R. Long noncoding RNA LINC00629 restrains the progression of gastric cancer by upregulating AQP4 through competitively binding to miR-196b-5p. J Cell Physiol. 2020;235:2973–2985. doi: 10.1002/jcp.29203. [DOI] [PubMed] [Google Scholar]
- 23.Than BL, Goos JA, Sarver AL, O’Sullivan MG, Rod A, Starr TK, Fijneman RJ, Meijer GA, Zhao L, Zhang Y, Largaespada DA, Scott PM, Cormier RT. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene. 2014;33:3861–3868. doi: 10.1038/onc.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.