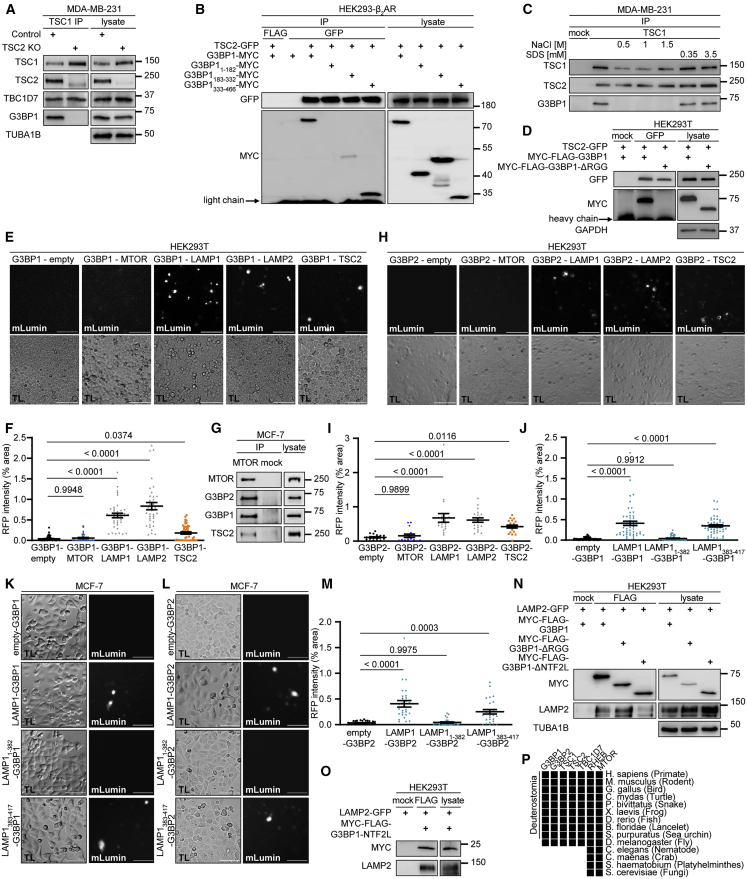
Figure 5.
G3BPs bridge TSC2 to LAMP proteins
(A) IP against TSC1 (TSC1 #1) or mock (rabbit IgG) in TSC2 KO cells. n = 3.
(B) IP against GFP or FLAG; transfection with the indicated plasmids. n = 5.
(C) IP against TSC1 (TSC1 #2) or mock (mouse IgG) incubated with NaCl or SDS. n = 3.
(D) IP against GFP or mock (mouse IgG); transfection with the indicated plasmids. n = 3.
(E) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 3.
(F) Quantitation of data in (E). Shown are data points and mean ± SEM.
(G) IP against MTOR or mock (rat IgG). n = 3.
(H) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 4.
(I) Quantitation of data in (H). Shown are data points and mean ± SEM.
(J) Quantitation of data in (K). Shown are data points and mean ± SEM.
(K) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 5.
(L) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 3.
(M) Quantitation of data in (L). Shown are data points and mean ± SEM.
(N) IP against FLAG or mock (mouse IgG); transfection with the indicated plasmids. n = 3.
(O) IP against FLAG or mock (mouse IgG); transfection with the indicated plasmids. n = 3.
(P) Phylogenetic analysis. Black square, protein present in species.
See also Figure S5.