Figure S6.
Background information for the zebrafish experiments, related to Figure 7
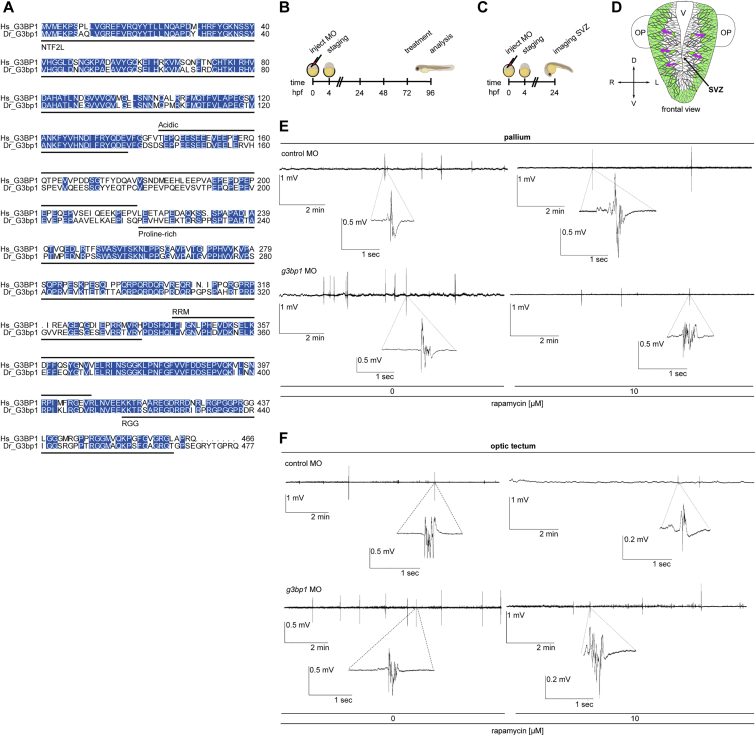
(A) Sequence alignment of human (Hs) G3BP1 (UniProt: Q13283) and zebrafish (Dr) G3bp1 (UniProt: Q6P124). Protein domains indicated according to Reineke and Lloyd (2015). Blue, identical residues. The sequences share 67.8% identity and 77.4% similarity.
(B) Treatment scheme of the g3bp1 MO and control MO injected zebrafish larvae for the analyses at 4 dpf (96 hpf): IF analysis, non-invasive local field potential (LFP) recordings, cell activity measurements, GABAergic and glutamatergic network analysis, locomotor tracking; treatment with rapamycin or ethosuximide at 3 dpf (72 hpf).
(C) Treatment scheme for the neuronal migration experiments with the Tg(HuC:GCaMP5G) transgenic line. Migration of HuC positive cells from the subventricular zone (SVZ) toward outer layers was analyzed at 24 hpf.
(D) Schematic frontal view of the zebrafish front brain at 24 hpf. Green, HuC expressing cells. Magenta arrows, direction of migration from the SVZ to outer layers. V, ventricle; OP, olfactory placodes.
(E) LFPs from larval pallia. Representative 10 min recordings of non-invasive LFPs from g3bp1 MO or control MO injected zebrafish larvae. Treatment as indicated in (B). Magnification of a polyspiking event is shown for each condition. n > 34 larvae per condition.
(F) LFPs from larval optic tecta. Representative 10 min recordings of non-invasive LFPs from g3bp1 MO or control MO injected zebrafish larvae. Treatment as indicated in (B). Magnification of a polyspiking event is shown for each condition. n > 20 larvae per condition.